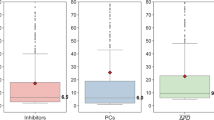
Abstract
Advances in increasingly complex phenotypic screening with lower throughput have necessitated the screening of smaller more highly annotated sets. One such collection of compounds which has been recently assembled is the kinase chemogenomic set. This is a set of curated kinase inhibitors built upon previous iterations, PKIS and PKIS2, and donations from our partners. Each compound in the set has been carefully selected based on selectivity, potency, and kinome coverage. These compounds as a set have been made available to the scientific community, enabling phenotypic screens to identify kinases that drive novel biology. Additionally, the associated data deposited in the public domain have also been used to inform new inhibitor design. Further expansion of this set to complete kinome coverage will allow for a greater understanding of kinase biology and its role in disease.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Jones LH, Bunnage ME (2017) Applications of chemogenomic library screening in drug discovery. Nat Rev Drug Discov 16(4):285–296
Canham SM, Wang Y, Cornett A, Auld DS, Baeschlin DK, Patoor M et al (2020) Systematic Chemogenetic library assembly. Cell Chem Biol 27(9):1124–1129
Gollner A, Koster M, Nicklin P, Trieselmann T, Klein E, Vlach J et al (2022) opnMe.com: a digital initiative for sharing tools with the biomedical research community. Nat Rev Drug Discov 21(7):475–476
Müller S, Ackloo S, Arrowsmith CH, Bauser M, Baryza JL, Blagg J et al (2018) Science Forum: donated chemical probes for open science. Elife 7:e34311
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298(5600):1912–1934
Wu P, Nielsen TE, Clausen MH (2015) FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci 36(7):422–439
Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J et al (2015) The promise and peril of chemical probes. Nat Chem Biol 11(8):536–541
Drewry DH, Wells CI, Andrews DM, Angell R, Al-Ali H, Axtman AD et al (2017) Progress towards a public chemogenomic set for protein kinases and a call for contributions. PLoS One 12(8):e0181585
Elkins JM, Fedele V, Szklarz M, Abdul Azeez KR, Salah E, Mikolajczyk J et al (2016) Comprehensive characterization of the published kinase inhibitor set. Nat Biotechnol 34(1):95–103
Bamborough P, Drewry D, Harper G, Smith GK, Schneider K (2008) Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem 51(24):7898–7914
Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G et al (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29(11):1046–1051
Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 29(11):1039–1045
Metz JT, Johnson EF, Soni NB, Merta PJ, Kifle L, Hajduk PJ (2011) Navigating the kinome. Nat Chem Biol 7(4):200–202
Gao Y, Davies SP, Augustin M, Woodward A, Patel UA, Kovelman R et al (2013) A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochem J 451(2):313–328
Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA et al (2017) The target landscape of clinical kinase drugs. Science 358(6367):eaan4368
Dranchak P, MacArthur R, Guha R, Zuercher WJ, Drewry DH, Auld DS et al (2013) Profile of the GSK published protein kinase inhibitor set across ATP-dependent and-independent luciferases: implications for reporter-gene assays. PLoS One 8(3):e57888
Khagi S, Sengupta S, Tabtieng T, Cochran BH (2015) Abstract C201: activity of the GlaxoSmithKline kinase inhibitor set on glioblastoma stem cells. Mol Cancer Ther 14(12_Supplement_2):C201-C
Wells C, Counago RM, Limas JC, Almeida TL, Cook JG, Drewry DH et al (2020) SGC-AAK1-1: a chemical probe targeting AAK1 and BMP2K. ACS Med Chem Lett 11(3):340–345
Asquith CRM, Berger BT, Wan J, Bennett JM, Capuzzi SJ, Crona DJ et al (2019) SGC-GAK-1: a chemical probe for cyclin G associated kinase (GAK). J Med Chem 62(5):2830–2836
Wells CI, Drewry DH, Pickett JE, Tjaden A, Kramer A, Muller S et al (2021) Development of a potent and selective chemical probe for the pleiotropic kinase CK2. Cell. Chem Biol 28(4):546–58 e10
Picado A, Chaikuad A, Wells CI, Shrestha S, Zuercher WJ, Pickett JE et al (2020) A chemical probe for dark kinase STK17B derives its potency and high selectivity through a unique P-loop conformation. J Med Chem 63(23):14626–14646
Wells CI, Al-Ali H, Andrews DM, Asquith CRM, Axtman AD, Dikic I et al (2021) The kinase Chemogenomic set (KCGS): an open science resource for kinase vulnerability identification. Int J Mol Sci 22(2):566
Deng X, Elkins JM, Zhang J, Yang Q, Erazo T, Gomez N et al (2013) Structural determinants for ERK5 (MAPK7) and leucine rich repeat kinase 2 activities of benzo[e]pyrimido-[5,4-b]diazepine-6(11H)-ones. Eur J Med Chem 70:758–767
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26
Schirmer AU, Driver LM, Zhao MT, Wells CI, Pickett JE, O’Bryne SN et al (2022) Non-canonical role of Hippo tumor suppressor serine/threonine kinase 3 STK3 in prostate cancer. Mol Ther 30(1):485–500
Vasta JD, Corona CR, Wilkinson J, Zimprich CA, Hartnett JR, Ingold MR et al (2018) Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem Biol 25(2):206–14. e11
Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB et al (2014) Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511(7511):616–620
Zhou W, Hur W, McDermott U, Dutt A, Xian W, Ficarro SB et al (2010) A structure-guided approach to creating covalent FGFR inhibitors. Chem Biol 17(3):285–295
Siddiqui-Jain A, Drygin D, Streiner N, Chua P, Pierre F, O’Brien SE et al (2010) CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res 70(24):10288–10298
Uitto J, McGrath JA, Rodeck U, Bruckner-Tuderman L, Robinson EC (2010) Progress in epidermolysis bullosa research: toward treatment and cure. J Invest Dermatol 130(7):1778–1784
Tan TS, Common JEA, Lim JSY, Badowski C, Firdaus MJ, Leonardi SS et al (2021) A cell-based drug discovery assay identifies inhibition of cell stress responses as a new approach to treatment of epidermolysis bullosa simplex. J Cell Sci 134(19):jcs258409
Schroder M, Filippakopoulos P, Schwalm MP, Ferrer CA, Drewry DH, Knapp S et al (2020) Crystal structure and inhibitor identifications reveal targeting opportunity for the atypical MAPK kinase ERK3. Int J Mol Sci 21(21):7953
Henderson SH, Sorrell F, Bennett J, Hanley MT, Robinson S, Hopkins Navratilova I et al (2020) Mining public domain data to develop selective DYRK1A inhibitors. ACS Med Chem Lett 11(8):1620–1626
Bao L, Wang Z, Wu Z, Luo H, Yu J, Kang Y et al (2023) Kinome-wide polypharmacology profiling of small molecules by multi-task graph isomorphism network approach. Acta Pharm Sin B 13(1):54–67
Wells CI, Vasta JD, Corona CR, Wilkinson J, Zimprich CA, Ingold MR et al (2020) Quantifying CDK inhibitor selectivity in live cells. Nat Commun 11(1):2743
Acknowledgement
This work was supported by the Eshelman Institute for Innovation (http://unceii.org/). The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome Trust [092809/Z/10/Z]. GlaxoSmithKline, Pfizer, Takeda, AstraZeneca, Boehringer Ingelheim, Bayer, Abbvie, Merck, and Nathanael Gray are gratefully acknowledged for donation of KCGS compounds.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Wells, C.I., Drewry, D.H. (2023). Developing a Kinase Chemogenomic Set: Facilitating Investigation into Kinase Biology by Linking Phenotypes to Targets. In: Merk, D., Chaikuad, A. (eds) Chemogenomics. Methods in Molecular Biology, vol 2706. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3397-7_2
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3397-7_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3396-0
Online ISBN: 978-1-0716-3397-7
eBook Packages: Springer Protocols