Abstract
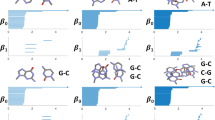
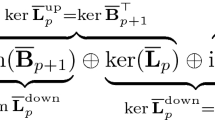
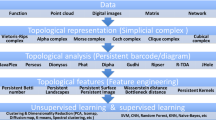
Molecular representations are of great importance for machine learning models in RNA data analysis. Essentially, efficient molecular descriptors or fingerprints that characterize the intrinsic structural and interactional information of RNAs can significantly boost the performance of all learning modeling. In this paper, we introduce two persistent models, including persistent homology and persistent spectral, for RNA structure and interaction representations and their applications in RNA data analysis. Different from traditional geometric and graph representations, persistent homology is built on simplicial complex, which is a generalization of graph models to higher-dimensional situations. Hypergraph is a further generalization of simplicial complexes and hypergraph-based embedded persistent homology has been proposed recently. Moreover, persistent spectral models, which combine filtration process with spectral models, including spectral graph, spectral simplicial complex, and spectral hypergraph, are proposed for molecular representation. The persistent attributes for RNAs can be obtained from these two persistent models and further combined with machine learning models for RNA structure, flexibility, dynamics, and function analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Singh J, Hanson J, Paliwal K, Zhou Y (2019) RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning.Nat Commun 10(1):1–13
Liu B (2019) BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief Bioinfor 20(4), 1280–1294
Puton T, Kozlowski LP, Rother KM, Bujnicki JM (2013) CompaRNA: a server for continuous benchmarking of automated methods for RNA secondary structure prediction. Nucleic Acids Res 41(7):4307–4323
Bellaousov S, Mathews DH (2010) ProbKnot: fast prediction of RNA secondary structure including pseudoknots. RNA 16(10):1870–1880
Guruge I, Taherzadeh G, Zhan J, Zhou Y, Yang Y (2018) B-factor profile prediction for RNA flexibility using support vector machines. J Comput Chem 39(8):407–411
Wei H, Wang B, Yang J, Gao J (2019) RNA flexibility prediction with sequence profile and predicted solvent accessibility. IEEE/ACM Trans Comput Biol Bioinf 18:2017–2022
Verri A, Uras C, Frosini P, Ferri M (1993) On the use of size functions for shape analysis. Biolog Cybern 70(2):99–107
Edelsbrunner H, Letscher D, Zomorodian A (2002) Topological persistence and simplification. Discrete Comput Geom 28:511–533
Zomorodian A, Carlsson G (2005) Computing persistent homology. Discrete Comput Geom 33:249–274
Zomorodian A, Carlsson G (2008) Localized homology. Comput Geom Theory Appl 41(3):126–148
Edelsbrunner H, Harer J (2010) Computational topology: an introduction. American Mathematical Society, Providence
Kaczynski T, Mischaikow K, Mrozek M (2004) Computational homology. Springer, Berlin
Xia KL, Wei GW (2014) Persistent homology analysis of protein structure, flexibility and folding. Int J Num Methods Biomed Eng 30:814–844
Wang B, Wei GW (2016) Object-oriented persistent homology. J Comput Phys 305:276–299
Cang ZX, Wei GW (2017) TopologyNet: topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLOS Comput Biol 13(7):e1005690
Cang ZX, Wei GW (2017) Integration of element specific persistent homology and machine learning for protein-ligand binding affinity prediction. Int J Numer Methods Biomed Eng 34:e2914. https://doi.org/10.1002/cnm.2914
Nguyen DD, Xiao T, Wang ML, Wei GW (2017) Rigidity strengthening: a mechanism for protein–ligand binding. J Chem Inf Modeling 57(7):1715–1721
Cang ZX, Wei GW (2017) Analysis and prediction of protein folding energy changes upon mutation by element specific persistent homology. Bioinformatics 33(22):3549–3557
Cang ZX, Mu L, Wei GW (2018) Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput Biol 14(1):e1005929
Wu KD, Wei GW (2018) Quantitative toxicity prediction using topology based multi-task deep neural networks. J Chem Inf Modeling 58:520–531. https://doi.org/10.1021/acs.jcim.7b00558
Ghrist R (2008) Barcodes: the persistent topology of data. Bull Amer Math Soc 45(1):61–75
Tausz A, Vejdemo-Johansson M, Adams H (2011) Javaplex: a research software package for persistent (co)homology. Software available at http://code.google.com/p/javaplex
Nanda V, Perseus: the persistent homology software. Software available at http://www.sas.upenn.edu/~vnanda/perseus
Bauer U, Kerber M, Reininghaus J (2014) Distributed computation of persistent homology. In: Proceedings of the sixteenth workshop on algorithm engineering and experiments (ALENEX)
Dionysus: the persistent homology software. Software available at http://www.mrzv.org/software/dionysus
Binchi J, Merelli E, Rucco M, Petri G, Vaccarino F (2014) jHoles: a tool for understanding biological complex networks via clique weight rank persistent homology. Electron Notes Theoret Comput Sci 306:5–18
Maria C (2015) Filtered complexes. In: GUDHI User and Reference Manual, GUDHI Editorial Board
Fasy BT, Kim J, Lecci F, Maria C (2014) Introduction to the R package TDA. Preprint arXiv:1411.1830
Mischaikow K, Nanda V (2013) Morse theory for filtrations and efficient computation of persistent homology. Discrete Comput Geom 50(2):330–353
Bubenik P, Kim PT (2007) A statistical approach to persistent homology. Homol Homotopy Appl 19:337–362
Bubenik P (2015) Statistical topological data analysis using persistence landscapes. J Mach Learn Res 16(1):77–102
Carlsson G (2009) Topology and data. Am Math Soc 46(2):255–308
Chintakunta H, Gentimis T, Gonzalez-Diaz R, Jimenez MJ, Krim H (2015) An entropy-based persistence barcode. Pattern Recogn 48(2):391–401
Merelli E, Rucco M, Sloot P, Tesei L (2015) Topological characterization of complex systems: Using persistent entropy. Entropy 17(10):6872–6892
Rucco M, Castiglione F, Merelli E, Pettini M (2016) Characterisation of the idiotypic immune network through persistent entropy. In: Proceedings of ECCS 2014, pp 117–128. Springer, Berlin
Xia KL, Li ZM, Mu L (2018) Multiscale persistent functions for biomolecular structure characterization. Bull Math Biol 80(1):1–31
Collins A, Zomorodian A, Carlsson G, Guibas LJ (2004) A barcode shape descriptor for curve point cloud data. Comput Graph 28(6):881–894
Cohen-Steiner D, Edelsbrunner H, Harer J (2007) Stability of persistence diagrams. Discrete Comput Geom 37(1):103–120
Cohen-Steiner D, Edelsbrunner H, Harer J, Mileyko Y (2010) Lipschitz functions have lp-stable persistence. Found Comput Math 10(2):127–139
Dawson RJM (1990) Homology of weighted simplicial complexes. Cahiers de Topologie et Géométrie Différentielle Catégoriques 31(3):229–243
Ren SQ, Wu CY, Wu J (2018) Weighted persistent homology. Rocky Mountain J Math 48(8):2661–2687
Wu CY, Ren SQ, Wu J, Xia KL (2018) Weighted (co) homology and weighted Laplacian. Sci China Math
Edelsbrunner H (1992) Weighted alpha shapes, vol 92. University of Illinois at Urbana-Champaign, Department of Computer Science, Champaign
Bell G, Lawson A, Martin J, Rudzinski J, Smyth C (2017) Weighted persistent homology. Preprint arXiv:1709.00097
Guibas L, Morozov D, Mérigot Q (2013) Witnessed k-distance. Discrete Comput Geom 49(1):22–45
Buchet M, Chazal F, Oudot SY, Sheehy DR (2016) Efficient and robust persistent homology for measures. Comput Geom 58:70–96
Xia KL, Wei GW (2015) Multidimensional persistence in biomolecular data. J Comput Chem 36:1502–1520
Xia KL, Zhao ZX, Wei GW (2015) Multiresolution persistent homology for excessively large biomolecular datasets. J Chem Phys 143(13):10B603_1
Petri G, Scolamiero M, Donato I, Vaccarino F (2013) Topological strata of weighted complex networks. PloS one 8(6):e66506
Xia KL, Wei GW (2014) Persistent homology analysis of protein structure, flexibility, and folding. Int J Numer Methods Biomed Eng 30(8):814–844
Nguyen DD, Cang ZX, Wu KD, Wang ML, Cao Y, Wei GW (2019) Mathematical deep learning for pose and binding affinity prediction and ranking in D3R Grand Challenges. J Comput-Aided Molec Design 33(1):71–82
Meng ZY, Anand DV, Lu YP, Wu J, Xia KL (2020) Weighted persistent homology for biomolecular data analysis. Sci Rep 10(1):1–15
Anand DV, Meng ZY, Xia KL, Mu YG (2020) Weighted persistent homology for osmolyte molecular aggregation and hydrogen-bonding network analysis. Sci Rep 10(1):1–17
Pun CS, Yong BYS, Xia K (2020) Weighted-persistent-homology-based machine learning for rna flexibility analysis. PloS one 15(8):e0237747
Chung F (1997) Spectral graph theory. American Mathematical Society, Providence
Spielman DA (2007) Spectral graph theory and its applications. In: 48th annual IEEE symposium on foundations of computer science (FOCS’07), pp 29–38, IEEE
Mohar B, Alavi Y, Chartrand G, Oellermann OR (1991) The Laplacian spectrum of graphs. Graph Theory Combin Appl 2(871–898):12
Von Luxburg U (2007) A tutorial on spectral clustering. Statist Comput 17(4):395–416
Eckmann B (1944) Harmonische funktionen und randwertaufgaben in einem komplex. Commen Math Helvetici 17(1):240–255
Muhammad A, Egerstedt M (2006) Control using higher order Laplacians in network topologies. In: Proceeding of the 17th international symposium on mathematical theory of networks and systems, pp 1024–1038. CiteSeer
Horak D, Jost J (2013) Spectra of combinatorial Laplace operators on simplicial complexes. Adv Math 244:303–336
Barbarossa S, Sardellitti S (2020) Topological signal processing over simplicial complexes. IEEE Trans Signal Process 68:2992–3007
Mukherjee S, Steenbergen J (2016) Random walks on simplicial complexes and harmonics. Random Struct Algor 49(2):379–405
Parzanchevski O, Rosenthal R (2017) Simplicial complexes: spectrum, homology and random walks. Random Struct Algor 50(2):225–261
Shukla S, Yogeshwaran D (2020) Spectral gap bounds for the simplicial Laplacian and an application to random complexes. J Combin Theory Ser A 169:105134
Torres JJ, Bianconi G (2020) Simplicial complexes: higher-order spectral dimension and dynamics. Preprint arXiv:2001.05934
Bramer D, Wei G-W (2018) Blind prediction of protein b-factor and flexibility. J Chem Phys 149(13):134107
Bramer D, Wei G-W (2020) Atom-specific persistent homology and its application to protein flexibility analysis. Comput Math Biophys 8(1):1–35
Wee J, Xia K (2021) Forman persistent Ricci curvature (FPRC) based machine learning models for protein-ligand binding affinity prediction. Briefings in Bioinformatics 22:bbab136
Wee J, Xia K (2021) Ollivier persistent Ricci curvature-based machine learning for the protein–ligand binding affinity prediction. J Chem Inf Modeling 61(4):1617–1626
Liu X, Wang XJ, Wu J, and Xia KL (2021) Hypergraph based persistent cohomology (HPC) for molecular representations in drug design. Briefings in Bioinformatics 22:bbaa411
Wang R, Nguyen DD, Wei G-W (2020) Persistent spectral graph. Int J Numer Methods Biomed Eng 36:e3376
Wang R, Zhao R, Ribando-Gros E, Chen J, Tong Y, Wei G-W (2020) HERMES: persistent spectral graph software. Found Data Sci 3:67–97
Zhao R, Wang M, Chen J, Tong Y, Wei G-W (2020) The de Rham-Hodge analysis and modeling of biomolecules. Bull Math Biol 82(8):1–38
Zhao R, Desbrun M, Wei G-W, Tong Y (2019) 3D Hodge decompositions of edge-and face-based vector fields. ACM Trans Graph (TOG) 38(6):1–13
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Xia, K., Liu, X., Wee, J. (2023). Persistent Homology for RNA Data Analysis. In: Filipek, S. (eds) Homology Modeling. Methods in Molecular Biology, vol 2627. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2974-1_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2974-1_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2973-4
Online ISBN: 978-1-0716-2974-1
eBook Packages: Springer Protocols