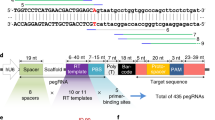
Abstract
Inherited retinal diseases (IRDs) encompass a large heterogeneous group of rare blinding disorders whose etiology originates from mutations in the 280 genes identified to date. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems represent a promising avenue for the treatment of IRDs, as exemplified by FDA clinical trial approval of EDIT-101 (AGN-151587), which removes a deep intronic variant in the CEP290 gene that causes Leber congenital amaurosis (LCA) type 10. Prime editing is a novel double-strand break (DSB) independent CRISPR/Cas system which has the potential to correct all 12 possible transition and transversion mutations in addition to small deletions and insertions. Here, as a proof-of-concept study, we describe a methodology using prime editing for the in vitro installation and correction of the classical Pde6brd10 c.1678C > T (p.Arg560Cys) mutation which causes autosomal recessive retinitis pigmentosa (RP) in mice.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Enoch J et al (2019) Evaluating whether sight is the most valued sense. JAMA Ophthalmol 137(11):1317–1320
Vu HT et al (2005) Impact of unilateral and bilateral vision loss on quality of life. Br J Ophthalmol 89(3):360–363
Maguire AM et al (2021) Clinical perspective: treating RPE65-associated retinal dystrophy. Mol Ther 29(2):442–463
Tsai YT et al (2018) Clustered regularly interspaced short palindromic repeats-based genome surgery for the treatment of autosomal dominant retinitis pigmentosa. Ophthalmology 125(9):1421–1430
Christie KA et al (2020) Mutation-independent allele-specific editing by CRISPR-Cas9, a novel approach to treat autosomal dominant disease. Mol Ther 28(8):1846–1857
Mussolino C et al (2011) AAV-mediated photoreceptor transduction of the pig cone-enriched retina. Gene Ther 18(7):637–645
Buck TM, Wijnholds J (2020) Recombinant adeno-associated viral vectors (rAAV)-vector elements in ocular gene therapy clinical trials and transgene expression and bioactivity assays. Int J Mol Sci 21(12)
Wu W et al (2021) Molecular and therapeutic strategies for retinitis pigmentosa: culture of human retinal explants for ex vivo assessment of AAV gene delivery, in methods in molecular biology, W. JM, editor. Humana Press, New York
Fry LE, McClements ME, MacLaren RE (2021) Analysis of pathogenic variants correctable with CRISPR base editing among patients with recessive inherited retinal degeneration. JAMA Ophthalmol 139(3):319–328
Tornabene P et al (2019) Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci Transl Med 11(492)
Gasiunas G et al (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109(39):E2579–E2586
Jinek M et al (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Mao Z et al (2008) DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7(18):2902–2906
Komor AC et al (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533(7603):420–424
Anzalone AV et al (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576(7785):149–157
Maeder ML et al (2019) Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med 25(2):229–233
Single ascending dose study in participants with LCA10. Allergan 2020; Available from: Editas Medicine, Inc.
Gallego C, Goncalves M, Wijnholds J (2020) Novel therapeutic approaches for the treatment of retinal degenerative diseases: focus on CRISPR/Cas-based gene editing. Front Neurosci 14:838
Vagni P et al (2019) Gene editing preserves visual functions in a mouse model of retinal degeneration. Front Neurosci 13:945
Chow RD et al (2021) A web tool for the design of prime-editing guide RNAs. Nat Biomed Eng 5(2):190–194
Hsu JY et al (2021) PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat Commun 12(1):1034
Clement K et al (2019) CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37(3):224–226
Park J et al (2017) Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33(2):286–288
Acknowledgement
B.L.D.C. is a recipient of the Capes PhD scholarship. S.H.T. and Jonas Children's Vision Care is supported by the National Institute of Health 5P30CA013696, U01 EY030580, U54OD020351, R24EY028758, R24EY027285, 5P30EY019007, R01EY018213, R01EY024698, R01EY026682, R21AG050437, the Schneeweiss Stem Cell Fund, New York State [SDHDOH01-C32590GG-3450000], the Foundation Fighting Blindness New York Regional Research Center Grant [TA-NMT-0116-0692-COLU], Nancy & Kobi Karp, the Crowley Family Funds, The Rosenbaum Family Foundation, Alcon Research Institute, the Gebroe Family Foundation, the Research to Prevent Blindness (RPB) Physician-Scientist Award, unrestricted funds from RPB, New York, NY, USA. P.M.J.Q. is the current recipient of a Curing Retinal Blindness Foundation (CRBF) grant, a Knights Templar Eye Foundation (KTEF) Career Starter grant, the International Retinal Research Foundation (IRRF) Loris and David Rich Postdoctoral Scholar Award and a New York Stem Cell Foundation (NYSCF)—Druckenmiller Fellowship.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Stephen H. Tsang receives financial support from Abeona Therapeutics, Inc and Emendo. He is also the founder of Rejuvitas and is on the scientific and clinical advisory board for Nanoscope Therapeutics. Peter M.J. Quinn receives research support from Rejuvitas, Inc.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Tsai, YT. et al. (2023). Prime Editing for the Installation and Correction of Mutations Causing Inherited Retinal Disease: A Brief Methodology. In: Tsang, S.H., Quinn, P.M. (eds) Retinitis Pigmentosa. Methods in Molecular Biology, vol 2560. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2651-1_29
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2651-1_29
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2650-4
Online ISBN: 978-1-0716-2651-1
eBook Packages: Springer Protocols