Abstract
High-throughput sequencing of adaptive immune receptor repertoires (AIRR, i.e., IG and TR) has revolutionized the ability to carry out large-scale experiments to study the adaptive immune response. Since the method was first introduced in 2009, AIRR sequencing (AIRR-Seq) has been applied to survey the immune state of individuals, identify antigen-specific or immune-state-associated signatures of immune responses, study the development of the antibody immune response, and guide the development of vaccines and antibody therapies. Recent advancements in the technology include sequencing at the single-cell level and in parallel with gene expression, which allows the introduction of multi-omics approaches to understand in detail the adaptive immune response. Analyzing AIRR-seq data can prove challenging even with high-quality sequencing, in part due to the many steps involved and the need to parameterize each step. In this chapter, we outline key factors to consider when preprocessing raw AIRR-Seq data and annotating the genetic origins of the rearranged receptors. We also highlight a number of common difficulties with common AIRR-seq data processing and provide strategies to address them.
Lmar Babrak and Susanna Marquez are shared first authors.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- AIRR-Seq
- B-cell receptor
- Germline database
- Gene annotation
- Preprocessing
- Single-cell sequencing
- T-cell receptor
1 Introduction
Once an Adaptive Immune Receptor Repertoire sequencing (AIRR-seq, please see the AIRR Community glossary at doi: https://doi.org/10.5281/zenodo.5095381 for definitions of key terms) experiment has been successfully designed and carried out (see discussion in the Chap. 15, attention turns to analyzing the data collected to produce biological insights. Many of the same factors that influenced choices in experimental design will be important in planning the computational approach as well. AIRR-seq data to be analyzed may have been generated from genomic DNA or mRNA, with or without unique molecular identifiers (UMIs), and in bulk or single-cell context, as described in the Chap. 15. Each of these alternatives may require (or preclude) the use of certain software tools and influence the interpretation of the analysis. In addition, thought must be given to what computational and storage resources will be necessary given the size of the dataset and the intended analysis.
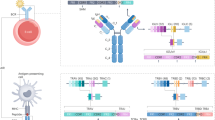
A clear first decision point in AIRR-seq data analysis is whether IG or TR repertoires are being analyzed (Fig. 1). While many tools such as MiXCR [1], IMGT [2], and others (Table 1) can handle both types of data, some are specific to one or the other. In addition, interest in specialized inquiries like phylogenetic analysis of IGs or calculation of clonal dynamics may require additional specific tools. In such a case, it may be useful to work within a particular ecosystem like Immcantation (http://immcantation.org), VDJServer [18], or SONAR [12], which provide several tools for a thorough analysis from quality control to clonal analysis, to facilitate smooth workflows.
The most critical set of considerations revolve around the origins of the molecules that were actually loaded into the sequencer (see Chap. 15). They may have been initially amplified from genomic DNA or from mRNA; the former results in exactly one initial copy of each productive V(D)J rearrangement in a cell, while the latter starts with several or many copies and may vary with cell type and activation state. When amplifying mRNA, the initial molecules may also be labeled with UMIs, which enable the correction of errors introduced by PCR and/or sequencing by identifying reads that are derived from the same original molecule. Of note, while the usage of UMIs enables experimental error correction, their usage necessitates a considerably larger sequencing depth due to consensus read building (for a more nuanced discussion, see, e.g., [20, 21]). UMIs may also be used when sequencing DNA, but that is currently less common in practice. UMIs can also be used to improve quantification, by collapsing apparent expansions due to differential amplification. Some specialized UMI protocols may also require particular matched software tools to fully utilize the advantages of those schemes [22]. Without UMIs, it is advisable to cluster highly similar reads to avoid overcounting, particularly for IG sequences, where errors and somatic hypermutation (SHM) are often indistinguishable.
It is also important to think about how molecules from the full repertoire get included into the pool to be amplified for sequencing. For mRNA-derived libraries, in particular, the efficiency of cDNA generation can be a significant bottleneck and may vary depending on the enzymes and protocol used in the reverse transcription (RT) reaction [23, 24]. The efficiency of the RT reaction can lead to a bias toward abundant species in the repertoire and concomitant dropout of rare ones. In addition, because of the diversity of V and J genes and their surrounding genetic context, many protocols use pools of primers to capture the full repertoire [25]. However, these primers may have different efficiencies in amplifying their respective targets, and some genes might be targeted by more than one primer in a pool. Other protocols circumvent this problem by adding 5′ anchors during reverse transcription [26]. In addition, IGs with high SHM can lose their ability to bind to an intended primer, resulting in the depletion of these sequences from the measured repertoire.
Recently, several high-throughput technologies have become popular for conducting AIRR-seq at single-cell resolution. These provide the most accurate, direct measurements of repertoire statistics and allow more biologically accurate definitions of clones. To do so, however, requires analysis tools that are capable of keeping heavy/light, alpha/beta, or gamma/delta chain sequences linked. The AIRR Community [27] (https://www.antibodysociety.org/the-airr-community/) is developing standardized representations for “receptors” and “cells” to facilitate these analyses and ensure data portability. In addition, single-cell IG and TR data can be easily linked to transcriptomic and other measurements for more comprehensive analyses.
The sequencing technology used must also be taken into account. Illumina paired-end sequencing requires an additional preprocessing step to reassemble the amplicon, and this may result in a bias against longer sequences, with less overlap between the two reads. Meanwhile, more error-prone long-read technologies require extra attention to quality control.
This chapter aims to guide bioinformaticians through the first steps in repertoire analysis, specifically the considerations and preparation of raw data for subsequent repertoire analysis (see Chap. 17). Firstly, this chapter provides in-depth information on the materials necessary to conduct the analysis, including computational resources for data preparation, available software tools, and germline database information (Fig. 2). The main portion of the chapter then discusses the considerations on data preprocessing and annotation of raw sequences with a reference germline database.
2 Materials
2.1 Computing Resources
AIRR-seq data are usually large and require specialized analysis methods and software tools. A typical Illumina MiSeq sequencing run generates 20–30 million 2 × 300 bp paired-end sequence reads which roughly corresponds to 15 GB of sequence data to be processed. Other platforms like NextSeq, which is useful in projects where the full V gene is not needed, creates about 400 million 2 × 150 bp paired-end reads. Because of the size of the datasets, the analysis can be computationally expensive, particularly the early analysis steps like preprocessing and gene annotation that process the majority of the sequence data. A standard desktop PC may take 3–5 days of constant processing for a single MiSeq run, so dedicated high-performance computational resources may be required. The institution may provide a cluster with high-performance computers for running analysis jobs. Commercial services like Amazon Web Services or Google Cloud can provide access to compute resources. However, this may come at added costs and could carry with them privacy concerns. Alternatively, there are free computing resources available. For AIRR-seq data, VDJServer provides free access to high-performance computing at the Texas Advanced Computing Center (TACC) through a graphical user interface [18]. VDJServer has also parallelized execution for tools such as IgBLAST, so more compute resources are utilized as the size of the input data grows. Analysis that takes days on a desktop PC might take only a few hours on VDJServer. An example workflow is provided in the AIRR Community Chap. 22 with instructions about using VDJServer for immune repertoire analysis.
2.2 Software Tools
Many tools are available for the first steps in AIRR-seq analysis [28,29,30,31]. Table 1 highlights several of the more commonly used programs. These are noted particularly because they support standardized AIRR data representations and are mostly free and open source, two key criteria among the AIRR software guidelines (https://docs.airr-community.org/en/stable/swtools/airr_swtools_standard.html). When deciding what are the right software tools to analyze data, besides computational requirements and expertise of the user, we recommend taking into consideration whether these tools use the AIRR Community standards and are AIRR-compliant. Tools that use the standard can easily be incorporated into complex workflows with other tools that share the same data format. Selecting AIRR-compliant software adds an additional layer of transparency to the analysis, because the source code is (1) available for inspection on a publicly available repository, (2) uses a versioning system, (3) has been tested, and (4) is available as a container (Docker, Singularity), among other quality requirements. The use of AIRR standards and of AIRR-compliant software supports the transparency, reproducibility, and rigor of research results.
2.3 Germline Databases
IG and TR germline databases are a requirement for accurate AIRR-seq analyses, regardless of the technique used (e.g., single cell vs. bulk). These databases guide the assignment of sequences to known and novel IG and TR genes/alleles, facilitating downstream sequence annotation and the accurate assessment of various repertoire features (e.g., gene/allele usage, SHM , clonal assignment, etc.; see AIRR Community Chaps. 18–20 for more detail). A germline database should ideally contain the most comprehensive and accurate set of possible IG /TR V, D, and J genes and alleles that best represent the genomic content of an organism. There are various sources of reference germline databases available, and occasionally a tool is limited by which database can be used for a particular analysis. Thus, the use of a particular database, or a combination of databases, may vary depending on the experimental objectives, as well as the particular species in which the AIRR-seq data has been generated. We therefore recommend investing effort in obtaining as accurate a database as possible. Table 2 describes currently available databases, focusing on those that are in active development.
IMGT [2] provides the most commonly used reference genome databases, but even for species of substantial research interest, these do not represent species diversity and can contain sequences reported in error [35, 36]. For TR genes and for IG genes from nonhuman species, however, few or no satisfactory alternatives exist. Ongoing initiatives seek to remedy this by continuously improving germline databases across species. Several programs are available to infer personalized databases from AIRR-seq data for each experimental subject (Table 1). VDJbase (https://www.vdjbase.org) is a resource that brings together AIRR-seq and genomic information to study population diversity and identify previously unreported alleles [34]. In 2019, the AIRR Community established the IARC (Inferred Allele Review Committee) to evaluate, document, and name human IGH alleles inferred from AIRR-seq data [37], and it is anticipated that this approach will be extended to other species and loci over time: The IARC’s work is supported and published by OGRDB (the Open Germline Receptor Database, https://ogrdb.airr-community.org), which provides full information regarding alleles, metadata on the repertoires from which they originated, and ref. 32.
3 Methods
Preprocessing and gene annotation of AIRR-seq data takes as input the sequencing files and returns a set of high-quality sequences for which V, D, and J allele calls can be made and structural elements can be identified. After further quality control filtering steps, a final set of sequences is selected and can be used to carry out more in-depth analyses (see Chap. 17). All steps should be carefully documented to maintain data provenance and allow the analysis to be reproduced; the AIRR Community has defined a set of MiAIRR data processing fields to standardize the representation of analysis steps [38]. Below, we outline the concepts involved in each phase of analysis and then supply detailed protocols, applying them to common use cases. We also provide further information on reporting and sharing AIRR-seq data.
3.1 Preprocessing
While there are several experimental technologies available for AIRR-seq studies from different experimental setups, most approaches typically produce the same raw data file format (.fastq) and share the ultimate goal of obtaining a final set of reads of high quality, particularly in the complementarity-determining region 3 (CDR3 ) region, representative of each B or T cell in the repertoire. The general steps that need to be performed include (1) filtering reads (e.g., removing PhiX spike-ins, short reads, and reads with a low Phred score or excessive ambiguous base calls), (2) identifying and removing primers and sequencing barcodes (if present), (3) building consensus sequences (using UMI or cell barcodes, if present), (4) merging mate pairs (if using a paired-end protocol), (5) masking low-quality positions, (6) annotating with constant (C) region (if present), and (7) collapsing duplicate sequences. For some of these steps, some considerations and adjustments need to be made depending on whether the data are from genomic DNA or RNA, B cells or T cells; bulk or single cell, paired or unpaired chains, and whether UMIs have been used (Fig. 1).
In the following we describe the important considerations to be made when preprocessing AIRR-seq samples.
3.1.1 Filtering by Sequence or by Clone
Current NGS methods introduce occasional base-call errors which may not be detectable from the associated quality scores. A common approach to avoid incorporating these sequences in downstream analyses is to threshold data based on the frequency of reads. This does not eliminate such errors but can reduce their influence on gross metrics of the underlying immune repertoire. To remove spurious sequences, a common approach taken, e.g., by MiXCR [1] and SONAR [12], is to collapse identical or near-identical sequences and drop those with fewer than a specified number of reads (usually two or three). This approach is preferred where individual sequences may be of low quality, for instance, if sequencing depth is low. However, this approach to filtering can result in nonuniform loss of data when libraries of different sequencing depths are compared. Alternatively, instead of a preprocessing step, all sequences passing quality control checks can be grouped into clones using the regular workflows described in the AIRR Community method Chaps. 18 and 19, and then clones that include fewer than the specified number of unique sequences are removed prior to downstream analysis. This may be appropriate for high-quality sequences, such as with UMIs and sufficient sequencing depth for robust error correction. Without this correction, errors in the CDR3 can lead to the inference of spurious clones.
3.1.2 Read Length-Related Effects
Long paired-end reads provide useful information for reliable V gene assignment as well as more comprehensive mapping of SHM in the case of IG gene rearrangements [39]. As read length increases, the quality of base calls degrades as sequences are generated, but paired-end sequencing allows for computational alignment of the overlapping regions. After alignment, sequencing errors at the ends of the sequences can be reduced as the higher-quality base call for each position that overlaps can be used. However, for longer sequences such as with RNA libraries capturing the constant region, the read length on the sequencer may need to be increased, reducing the overlapping portion of the 5′ and 3′ reads, resulting in a bias against sequences encoding longer CDR3 . Further complicating this issue, a common procedure is to trim the ends of reads of low-quality stretches of base calls, such as with generic tools like fastx-toolkit or pRESTO’s FilterSeq trimqual [4]. This can in turn reduce the number of full-length high-quality sequences. On the other hand, with RNA-based sequencing, UMIs can be incorporated at the cDNA synthesis step, and, when coupled with very deep sequencing, these can be used for error correction through the construction of consensus sequences that share the same UMI . There is, however, a trade-off between the sequencing depth required for adequate coverage of UMIs and the number of independent sequences that can be sampled.
Long reads covering the entire variable region can also be generated using alternative sequencing platforms, such as those offered by Pacific Biosciences and Ion Torrent [31, 40,41,42,43]. These offer the additional advantage of being able to capture large enough parts of the C-region to be able to distinguish between subtypes of IgG. However, lower throughput on these platforms limits the depth of sampling that can be achieved.
Short reads are sometimes used to generate large quantities of data on CDR3 sequences, as sequencing short reads can be done on higher-throughput sequencers at lower cost. This strategy is particularly common for TR rearrangement analysis on gDNA using commercial platforms such as Adaptive. Short reads may be required if the template is of low quality, as sometimes occurs in formalin-fixed paraffin-embedded samples. Short reads can sometimes compromise TRBV gene assignments but are particularly problematic for IGH gene rearrangements with SHM . Short IGHV gene sequences result in larger numbers of ambiguous V gene assignments which can cause erroneous clustering of unrelated sequences into clones.
gDNA vs. mRNA templates. When using genomic DNA as starting material, each cell contributes a fixed number of IG or TR template, providing a parsimonious and cost-effective means of profiling large numbers of cells. gDNA-based sequencing will also capture far more nonproductive gene rearrangements than mRNA-based sequencing. With RNA, nonproductive rearrangements are subjected to nonsense-mediated degradation (although some nonproductive rearrangements can be recovered). gDNA is also more stable than RNA. On the other hand, RNA-based sequencing is more sensitive, with more templates per cell. With mRNA-based sequencing, cells contribute different numbers of templates, based upon cell subset-specific differences in transcript abundance. With mRNA-based libraries, cells can be grouped into subsets using immunophenotyping or single-cell RNA-seq to control for these differences. In the case of IG data where primers can be designed to capture the C-regions, each read can be annotated with its isotype using, for example, pRESTO’s MaskPrimers routine. Further, unlike gDNA, it is straightforward to incorporate unique molecular identifiers (UMIs) at the RNA to cDNA synthesis step. Each UMI , which should be unique to original individual cDNA templates, can be processed with pRESTO’s BuildConsensus to generate consensus sequences which can nearly eliminate sequencing error given sufficient sequencing depth [44, 45]. MiXCR, SONAR, and other packages also offer similar tools. The necessary depth might be difficult to achieve, though, for instance, in cases of vastly different expression levels or with samples of large size.
3.1.3 Productive Vs. Nonproductive Rearrangements
For each sample, the fraction of productive rearrangements can be an informative metric. On average, it can be expected that approximately 80% of TRB rearrangements and approximately 85% of IGH rearrangements sequenced from mature T or B cells will be productive [46]. Lower frequencies of productive rearrangements can be observed in immature lymphocytes, where selection has not yet been imposed on cells without productive rearrangements [47]. Lower frequencies of productive rearrangements can also be seen in sequencing libraries that are of poor quality. Nonproductive sequences also can be used as a baseline estimator of gene usage frequency in rearrangement [48, 49] and compared to productive sequences to investigate the effects of tolerance checkpoints on the AIRR [50, 51]. With such comparisons, it may be useful to remove clonal lineages that contain both productive and nonproductive versions of the same rearrangement, as sequencing errors can cause a sequence to appear nonproductive. Nonproductive rearrangements are sometimes also useful for identifying clonal expansions in tumors, particularly if tumors harbor SHM that may interfere with primer binding (the nonproductive rearrangements are usually un-mutated). Nonproductive rearrangements can be found in lymphocytes that have undergone multiple rounds of V(D)J recombination, as can occur with receptor editing; the presence of more than one rearrangement is particularly common with IG light chains [52, 53]. Finally, it is important to computationally filter nonproductive sequences for general analyses, if one is making claims about selected repertoires.
3.2 Gene Annotation
After preprocessing AIRR sequences for good-quality and relevant reads, sequences need to be accurately aligned and annotated to an appropriate reference germline database. This process identifies the V, D, and J genes; CDRs; and framework regions (FWRs) for each sequence in the repertoire. There are numerous annotation tools for IG and TR sequences that are freely available to users, including popular programs such as IgBLAST [10] and IMGT/HighV-QUEST (Table 1) [8]. Depending on the tools, different tool-specific algorithms (e.g., Smith-Waterman) assign the best match among a set of genes in a user-defined reference germline database. Accurate alignment is very important for subsequent analyses such as the identification of SHM for IGs, clustering of clonal groups, and determination of IG /TR diversity. Alignment algorithms have been demonstrated to influence the outcome of V, D, and J gene assignments, even when identical input sequences, tool parameters, and reference germline databases are chosen [31]. Furthermore, differences in the length of alleles of genes in databases may force algorithms to output an incorrect best match in the gene annotation process. To complicate matters, some tools provide alignments to multiple (often highly similar) genes and leave it to users to choose which of the ambiguous calls is most appropriate.
Schemes for IGs and TRs that number amino acid residues facilitate sequence comparisons, protein structure modelling, and engineering [54]. Although many schemes have been proposed and different schemes are employed by different tools, only five schemes are commonly used. Three are specifically for IGs: Kabat [55], Chothia [56], and enhanced Chothia [57]. Two more can be used for both IGs and TRs: IMGT [58] and AHo [59]. Conversion tables and tools like ANARCI [60] can be used to translate between schemes. CDR boundaries can differ substantially between different numbering schemes: care is needed when comparing results from different studies [54]. In repertoire studies, the IMGT numbering scheme is widely used and supported, and its use is recommended in the absence of other considerations.
One more barrier to direct comparison is the identification in some studies and tools of the “junction” and in others of the CDR3 . In IMGT terminology, the junction includes the second conserved cysteine of the V gene and the conserved tryptophan or phenylalanine of the J gene, while the CDR3 omits these residues. The AIRR Community data representation standard uses “junction”; however, it is not universally accepted [31].
Accurate annotation requires an accurate and comprehensive germline database. As noted above, even the currently available human database does not as yet meet this criterion [15, 61], and databases for other species are often partial and based solely on the analysis of a single animal [36, 62,63,64,65]. Fortunately, scientific need has resulted in the determination of new germline gene sets [36, 40, 66, 67], but these are not necessarily implemented by public germline gene databases in a timely fashion. The impact of missing or incorrect information in the database will depend upon the nature of the analysis, but one overall point to note is that the databases are updated frequently, and changes in the database can impact results [31]. It is therefore important that an analysis is conducted using a single, consistent, and up-to-date version of the database and that the version (or download date) is recorded for reproducibility. Germline databases are sometimes installed automatically with annotation tools: where that is the case, researchers should check if the installed version meets these requirements, and update it if necessary.
In a repertoire from a single individual, although structural variation and gene duplication give rise to frequent exceptions, we would expect to see a maximum of two alleles of most germline receptor genes: one from the paternal and one from the maternal chromosome. When used with an extensive germline database, annotation tools that are based on sequence similarity tend to call a biologically implausible number of alleles in B-cell repertoires, particularly in repertoires that are highly mutated, and will make a large number of indeterminate calls, where the tool would be unable to determine the likely germline allele unambiguously. Tools are available that will improve allele calls by using probabilistic methods to infer the individual’s “personalized” germline set: such tools can also infer the presence of alleles in the individual that were not listed in the annotation tool’s germline database [15,16,17, 68, 69]. While the use of a comprehensive germline database is important in the first instance, the determination of a personalized germline set and re-annotation with just that set is recommended where allele assignment is important: for example, when clonal inference is employed: personalization can also compensate to some extent for deficiencies in the germline database.
The decision of which annotation tool to use is also dependent on the computer skill set of the user. IMGT/HIGHV-QUEST and IgBLAST provide easy-to-use web platforms, suited for researchers that prefer to access a graphic user interface. Other tools, such as the stand-alone version of IgBLAST [10], MiXCR [1], and partis [11], require additional computer expertise, because they need to be installed and are used in the terminal. The advantage of such tools is that they provide more flexibility and can be integrated in automated workflows.
4 Conclusion
In this chapter, we present important considerations involved in the first steps in the preparation of raw data after sequencing and guide bioinformaticians in choosing the appropriate parameters for preprocessing and annotation. These first steps are required for the subsequent repertoire analysis, described in the Chap. 17, as choices made in these first steps have serious implications for the types of data analyses that can be performed and for the accuracy of the results. After the completion of this chapter, the bioinformatician is now ready to begin the in-depth analysis of repertoire features specific to the question at hand.
References
Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV et al (2015) MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 12:380–381. https://doi.org/10.1038/nmeth.3364
Giudicelli V, Brochet X, Lefranc M-P (2011) IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011:695–715. https://doi.org/10.1101/pdb.prot5633
Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH (2015) Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31:3356–3358. https://doi.org/10.1093/bioinformatics/btv359
Vander Heiden JA, Yaari G, Uduman M, Stern JNH, O’Connor KC, Hafler DA et al (2014) pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30:1930–1932. https://doi.org/10.1093/bioinformatics/btu138
Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G et al (2016) T cell fate and clonality inference from single-cell transcriptomes. Nat Methods 13:329–332. https://doi.org/10.1038/nmeth.3800
Christley S, Levin MK, Toby IT, Fonner JM, Monson NL, Rounds WH et al (2017) VDJPipe: a pipelined tool for pre-processing immune repertoire sequencing data. BMC Bioinformatics 18:448. https://doi.org/10.1186/s12859-017-1853-z
Peacock T, Heather JM, Ronel T, Chain B (2020) Decombinator V4 - an improved AIRR-compliant software package for T cell receptor sequence annotation. Bioinformatics 37(6):876–878. https://doi.org/10.1093/bioinformatics/btaa758
Alamyar E, Duroux P, Lefranc M-P, Giudicelli V (2012) IMGT® tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. In: Christiansen FT, Tait BD (eds) Immunogenetics. Humana Press, Totowa, NJ, pp 569–604. https://doi.org/10.1007/978-1-61779-842-9_32
Kuchenbecker L, Nienen M, Hecht J, Neumann AU, Babel N, Reinert K et al (2015) IMSEQ—a fast and error aware approach to immunogenetic sequence analysis. Bioinformatics 31:2963–2971. https://doi.org/10.1093/bioinformatics/btv309
Ye J, Ma N, Madden TL, Ostell JM (2013) IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 41:W34–W40. https://doi.org/10.1093/nar/gkt382
Ralph DK, Matsen FA (2016) Consistency of VDJ rearrangement and substitution parameters enables accurate B cell receptor sequence annotation. PLoS Comput Biol 12:e1004409. https://doi.org/10.1371/journal.pcbi.1004409
Schramm CA, Sheng Z, Zhang Z, Mascola JR, Kwong PD, Shapiro L (2016) SONAR: a high-throughput pipeline for inferring antibody ontogenies from longitudinal sequencing of B cell transcripts. Front Immunol 7:372. https://doi.org/10.3389/fimmu.2016.00372
Giraud M, Salson M, Duez M, Villenet C, Quief S, Caillault A et al (2014) Fast multiclonal clusterization of V(D)J recombinations from high-throughput sequencing. BMC Genomics 15:409. https://doi.org/10.1186/1471-2164-15-409
Duez M, Giraud M, Herbert R, Rocher T, Salson M, Thonier F (2016) Vidjil: a web platform for analysis of high-throughput repertoire sequencing. PLoS One 11:e0166126. https://doi.org/10.1371/journal.pone.0166126
Gadala-Maria D, Yaari G, Uduman M, Kleinstein SH (2015) Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc Natl Acad Sci U S A 112:E862–E870. https://doi.org/10.1073/pnas.1417683112
Ralph DK, Matsen FA (2019) Per-sample immunoglobulin germline inference from B cell receptor deep sequencing data. PLoS Comput Biol 15:e1007133. https://doi.org/10.1371/journal.pcbi.1007133
Corcoran MM, Phad GE, Bernat NV, Stahl-Hennig C, Sumida N, Persson MAA et al (2016) Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 7:13642. https://doi.org/10.1038/ncomms13642
Christley S, Scarborough W, Salinas E, Rounds WH, Toby IT, Fonner JM et al (2018) VDJServer: a cloud-based analysis portal and data commons for immune repertoire sequences and rearrangements. Front Immunol 9:976. https://doi.org/10.3389/fimmu.2018.00976
Rosenfeld AM, Meng W, Luning Prak ET, Hershberg U (2018) ImmuneDB, a novel tool for the analysis, storage, and dissemination of immune repertoire sequencing data. Front Immunol 9:2107. https://doi.org/10.3389/fimmu.2018.02107
Barennes P, Quiniou V, Shugay M, Egorov ES, Davydov AN, Chudakov DM et al (2021) Benchmarking of T cell receptor repertoire profiling methods reveals large systematic biases. Nat Biotechnol 39:236–245. https://doi.org/10.1038/s41587-020-0656-3
Greiff V, Miho E, Menzel U, Reddy ST (2015) Bioinformatic and statistical analysis of adaptive immune repertoires. Trends Immunol 36:738–749. https://doi.org/10.1016/j.it.2015.09.006
Khan TA, Friedensohn S, Gorter de Vries AR, Straszewski J, Ruscheweyh H-J, Reddy ST (2016) Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv 2:e1501371. https://doi.org/10.1126/sciadv.1501371
Schwaber J, Andersen S, Nielsen L (2019) Shedding light: the importance of reverse transcription efficiency standards in data interpretation. Biomol Detect Quantif 17:100077. https://doi.org/10.1016/j.bdq.2018.12.002
Zucha D, Androvic P, Kubista M, Valihrach L (2020) Performance comparison of reverse transcriptases for single-cell studies. Clin Chem 66:217–228. https://doi.org/10.1373/clinchem.2019.307835
van Dongen JJM, Langerak AW, Brüggemann M, Evans PAS, Hummel M, Lavender FL et al (2003) Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia 17:2257–2317. https://doi.org/10.1038/sj.leu.2403202
Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai K-L et al (2002) A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol 168:3099–3104. https://doi.org/10.4049/jimmunol.168.6.3099
Breden F, Luning Prak ET, Peters B, Rubelt F, Schramm CA, Busse CE et al (2017) Reproducibility and reuse of adaptive immune receptor repertoire data. Front Immunol 8:1418. https://doi.org/10.3389/fimmu.2017.01418
Zhang Y, Yang X, Zhang Y, Zhang Y, Wang M, Ou JX et al (2020) Tools for fundamental analysis functions of TCR repertoires: a systematic comparison. Brief Bioinform 21:1706–1716. https://doi.org/10.1093/bib/bbz092
López-Santibáñez-Jácome L, Avendaño-Vázquez SE, Flores-Jasso CF (2019) The pipeline repertoire for Ig-seq analysis. Front Immunol 10:899. https://doi.org/10.3389/fimmu.2019.00899
Lees WD (2020) Tools for adaptive immune receptor repertoire sequencing. Curr Opin Syst Biol 24:86–92. https://doi.org/10.1016/j.coisb.2020.10.003
Smakaj E, Babrak L, Ohlin M, Shugay M, Briney B, Tosoni D et al (2020) Benchmarking immunoinformatic tools for the analysis of antibody repertoire sequences. Bioinformatics 36:1731–1739. https://doi.org/10.1093/bioinformatics/btz845
Lees W, Busse CE, Corcoran M, Ohlin M, Scheepers C, Matsen FA et al (2020) OGRDB: a reference database of inferred immune receptor genes. Nucleic Acids Res 48:D964–D970. https://doi.org/10.1093/nar/gkz822
Giudicelli V, Chaume D, Lefranc M-P (2005) IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res 33:D256–D261. https://doi.org/10.1093/nar/gki010
Omer A, Shemesh O, Peres A, Polak P, Shepherd AJ, Watson CT et al (2020) VDJbase: an adaptive immune receptor genotype and haplotype database. Nucleic Acids Res 48:D1051–D1056. https://doi.org/10.1093/nar/gkz872
Wang Y, Jackson KJL, Sewell WA, Collins AM (2008) Many human immunoglobulin heavy-chain IGHV gene polymorphisms have been reported in error. Immunol Cell Biol 86:111–115. https://doi.org/10.1038/sj.icb.7100144
Vázquez Bernat N, Corcoran M, Nowak I, Kaduk M, Castro Dopico X, Narang S et al (2021) Rhesus and cynomolgus macaque immunoglobulin heavy-chain genotyping yields comprehensive databases of germline VDJ alleles. Immunity 54:355–366.e4. https://doi.org/10.1016/j.immuni.2020.12.018
Ohlin M, Scheepers C, Corcoran M, Lees WD, Busse CE, Bagnara D et al (2019) Inferred allelic variants of immunoglobulin receptor genes: a system for their evaluation, documentation, and naming. Front Immunol 10:435. https://doi.org/10.3389/fimmu.2019.00435
Rubelt F, Busse CE, Bukhari SAC, Bürckert J-P, Mariotti-Ferrandiz E, Cowell LG et al (2017) Adaptive immune receptor repertoire community recommendations for sharing immune-repertoire sequencing data. Nat Immunol 18:1274–1278. https://doi.org/10.1038/ni.3873
Zhang B, Meng W, Luning Prak ET, Hershberg U (2015) Discrimination of germline V genes at different sequencing lengths and mutational burdens: a new tool for identifying and evaluating the reliability of V gene assignment. J Immunol Methods 427:105–116. https://doi.org/10.1016/j.jim.2015.10.009
Watson CT, Kos JT, Gibson WS, Newman L, Deikus G, Busse CE et al (2019) A comparison of immunoglobulin IGHV, IGHD and IGHJ genes in wild-derived and classical inbred mouse strains. Immunol Cell Biol 97:888–901. https://doi.org/10.1111/imcb.12288
Deiss TC, Vadnais M, Wang F, Chen PL, Torkamani A, Mwangi W et al (2019) Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. Cell Mol Immunol 16:53–64. https://doi.org/10.1038/cmi.2017.117
Koning MT, Kiełbasa SM, Boersma V, Buermans HPJ, van der Zeeuw SAJ, van Bergen CAM et al (2017) ARTISAN PCR: rapid identification of full-length immunoglobulin rearrangements without primer binding bias. Br J Haematol 178:983–986. https://doi.org/10.1111/bjh.14180
Lay L, Stroup B, Payton JE (2020) Validation and interpretation of IGH and TCR clonality testing by ion torrent S5 NGS for diagnosis and disease monitoring in B and T cell cancers. Pract Lab Med 22:e00191. https://doi.org/10.1016/j.plabm.2020.e00191
Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B (2011) Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 108:9530–9535. https://doi.org/10.1073/pnas.1105422108
Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR et al (2014) Towards error-free profiling of immune repertoires. Nat Methods 11:653–655. https://doi.org/10.1038/nmeth.2960
Rubelt F, Bolen CR, McGuire HM, Heiden JAV, Gadala-Maria D, Levin M et al (2016) Individual heritable differences result in unique cell lymphocyte receptor repertoires of naïve and antigen-experienced cells. Nat Commun 7:11112. https://doi.org/10.1038/ncomms11112
Meng W, Yunk L, Wang L-S, Maganty A, Xue E, Cohen PL et al (2011) Selection of individual VH genes occurs at the pro-B to pre-B cell transition. J Immunol 187:1835–1844. https://doi.org/10.4049/jimmunol.1100207
Marcou Q, Mora T, Walczak AM (2018) High-throughput immune repertoire analysis with IGoR. Nat Commun 9:561. https://doi.org/10.1038/s41467-018-02832-w
Sethna Z, Elhanati Y, Callan CG, Walczak AM, Mora T (2019) OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinformatics 35:2974–2981. https://doi.org/10.1093/bioinformatics/btz035
Elhanati Y, Sethna Z, Marcou Q, Callan CG, Mora T, Walczak AM (2015) Inferring processes underlying B-cell repertoire diversity. Philos Trans R Soc Lond Ser B Biol Sci 370:20140243. https://doi.org/10.1098/rstb.2014.0243
Sethna Z, Isacchini G, Dupic T, Mora T, Walczak AM, Elhanati Y (2020) Population variability in the generation and selection of T-cell repertoires. PLoS Comput Biol 16:e1008394. https://doi.org/10.1371/journal.pcbi.1008394
Langerak AW, van Dongen JJM (2012) Multiple clonal Ig/TCR products: implications for interpretation of clonality findings. J Hematop 5:35–43. https://doi.org/10.1007/s12308-011-0129-1
Luning Prak ET, Monestier M, Eisenberg RA (2011) B cell receptor editing in tolerance and autoimmunity. Ann N Y Acad Sci 1217:96–121. https://doi.org/10.1111/j.1749-6632.2010.05877.x
Dondelinger M, Filée P, Sauvage E, Quinting B, Muyldermans S, Galleni M et al (2018) Understanding the significance and implications of antibody numbering and antigen-binding surface/residue definition. Front Immunol 9:2278. https://doi.org/10.3389/fimmu.2018.02278
Wu TT, Kabat EA (1970) An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med 132:211–250. https://doi.org/10.1084/jem.132.2.211
Al-Lazikani B, Lesk AM, Chothia C (1997) Standard conformations for the canonical structures of immunoglobulins. J Mol Biol 273:927–948. https://doi.org/10.1006/jmbi.1997.1354
Abhinandan KR, Martin ACR (2010) Analysis and prediction of VH/VL packing in antibodies. Protein Eng Des Sel 23:689–697. https://doi.org/10.1093/protein/gzq043
Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L et al (2003) IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 27:55–77. https://doi.org/10.1016/s0145-305x(02)00039-3
Honegger A, Plückthun A (2001) Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol 309:657–670. https://doi.org/10.1006/jmbi.2001.4662
Dunbar J, Deane CM (2016) ANARCI: antigen receptor numbering and receptor classification. Bioinformatics 32:298–300. https://doi.org/10.1093/bioinformatics/btv552
Watson CT, Breden F (2012) The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun 13:363–373. https://doi.org/10.1038/gene.2012.12
Ramesh A, Darko S, Hua A, Overman G, Ransier A, Francica JR et al (2017) Structure and diversity of the rhesus macaque immunoglobulin loci through multiple de novo genome assemblies. Front Immunol 8:1407. https://doi.org/10.3389/fimmu.2017.01407
Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA et al (2019) Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell 177:1153–1171.e28. https://doi.org/10.1016/j.cell.2019.04.012
Retter I, Chevillard C, Scharfe M, Conrad A, Hafner M, Im T-H et al (2007) Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J Immunol 179:2419–2427. https://doi.org/10.4049/jimmunol.179.4.2419
Collins AM, Wang Y, Roskin KM, Marquis CP, Jackson KJL (2015) The mouse antibody heavy chain repertoire is germline-focused and highly variable between inbred strains. Philos Trans R Soc Lond Ser B Biol Sci 370:20140236. https://doi.org/10.1098/rstb.2014.0236
Magadan S, Krasnov A, Hadi-Saljoqi S, Afanasyev S, Mondot S, Lallias D et al (2019) Standardized IMGT® nomenclature of Salmonidae IGH genes, the paradigm of Atlantic Salmon and rainbow trout: from genomics to repertoires. Front Immunol 10:2541. https://doi.org/10.3389/fimmu.2019.02541
Magadan S, Mondot S, Palti Y, Gao G, Lefranc MP, Boudinot P (2021) Genomic analysis of a second rainbow trout line (Arlee) leads to an extended description of the IGH VDJ gene repertoire. Dev Comp Immunol 118:103998. https://doi.org/10.1016/j.dci.2021.103998
Zhang W, Wang I-M, Wang C, Lin L, Chai X, Wu J et al (2016) IMPre: an accurate and efficient software for prediction of T- and B-cell receptor germline genes and alleles from rearranged repertoire data. Front Immunol 7:457. https://doi.org/10.3389/fimmu.2016.00457
Gadala-Maria D, Gidoni M, Marquez S, Vander Heiden JA, Kos JT, Watson CT et al (2019) Identification of subject-specific immunoglobulin alleles from expressed repertoire sequencing data. Front Immunol 10:129. https://doi.org/10.3389/fimmu.2019.00129
Acknowledgments
The authors would like to thank Eline T. Luning Prak for the constructive criticism of the manuscript.
Author information
Authors and Affiliations
Consortia
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Babrak, L. et al. (2022). Adaptive Immune Receptor Repertoire (AIRR) Community Guide to TR and IG Gene Annotation. In: Langerak, A.W. (eds) Immunogenetics. Methods in Molecular Biology, vol 2453. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2115-8_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2115-8_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2114-1
Online ISBN: 978-1-0716-2115-8
eBook Packages: Springer Protocols