Abstract
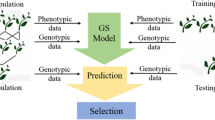
Continued improvement and falling costs of DNA sequencing have accelerated the increase in genomic resources for crop plants. From these efforts, considerable genetic diversity has been found and is aiding in the identification of markers for breeding purposes. High-density molecular markers have allowed for marker-assisted selection of quantitative traits that are controlled by a small number of genes. Recently, whole genomic selection has been proposed where markers genome-wide are used to estimate the contribution of all loci to traits of interest. In this chapter we outline the steps needed to perform genomic selection using machine learning. We describe our method called Crop Genomic Breeding Machine (CropGBM) and demonstrate its use on diverse maize lines containing high-density markers.
Software website: https://ibreeding.github.io/
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124(3):743–756
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664. https://doi.org/10.2135/cropsci2008.03.0131
Xu Y, Crouch J (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48:391–407. https://doi.org/10.2135/cropsci2007.04.0191
Heffner E, Sorrells M, Jannink J-L (2009) Genomic selection for crop improvement. Crop Sci 49:1–12. https://doi.org/10.2135/cropsci2008.08.0512
Meuwissen THE, Hayes BJB, Goddard MEM (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Xu S, Zhu D, Zhang Q (2014) Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc Natl Acad Sci U S A 111:12456–12461. https://doi.org/10.1073/pnas.1413750111
Kadam DC, Potts SM, Bohn MO, Lipka AE, Lorenz AJ (2016) Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3 (Bethesda, Md) 6(11):3443–3453. https://doi.org/10.1534/g3.116.031286
Henderson CR (1984) Applications of linear models in animal breeding. University of Guelph, Guelph
Bernardo R, Yu J (2007) Prospects for genomewide selection for quantitative traits in maize. Crop Sci 47:1082–1090. https://doi.org/10.2135/cropsci2006.11.0690
Maros M, Capper D, Jones D, Hovestadt V, Deimling A, Pfister S, Benner A, Zucknick M, Sill M (2020) Machine learning workflows to estimate class probabilities for precision cancer diagnostics on DNA methylation microarray data. Nat Protoc 15:479–512. https://doi.org/10.1038/s41596-019-0251-6
Dorman SN, Baranova K, Knoll JHM, Urquhart Brad L, Mariani G, Carcangiu ML, Rogan Peter K (2016) Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Mol Oncol 10(1):85–100. https://doi.org/10.1016/j.molonc.2015.07.006
Qiu Z, Cheng Q, Song J, Tang Y, Ma C (2016) Application of machine learning-based classification to genomic selection and performance improvement, vol 9771. doi:https://doi.org/10.1007/978-3-319-42291-6_41
Shakoor N, Lee S, Mockler T (2017) High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr Opin Plant Biol 38:184–192. https://doi.org/10.1016/j.pbi.2017.05.006
Acknowledgments
This work was supported by the National Science Foundation of China (31871706): Utilization of Machine Learning Strategy to Predict Maize Heterosis.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Xu, Y., Laurie, J.D., Wang, X. (2022). CropGBM: An Ultra-Efficient Machine Learning Toolbox for Genomic Selection-Assisted Breeding in Crops. In: Bilichak, A., Laurie, J.D. (eds) Accelerated Breeding of Cereal Crops. Springer Protocols Handbooks. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1526-3_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1526-3_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1525-6
Online ISBN: 978-1-0716-1526-3
eBook Packages: Springer Protocols