Abstract
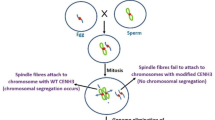
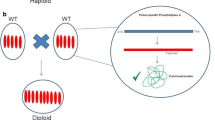
Haploid production is of great importance in plant breeding programs. Doubled haploid technology accelerates the generation of inbred lines with homozygosity in all loci in a single year. Haploids can be induced in vitro via cultivating the haploid gametes or in vivo through inter- and intraspecific hybridization. Haploid induction through centromere engineering is a novel system that is theoretically applicable to many plant species. The present review chapter discusses the proposed molecular mechanisms of selective chromosome elimination in early embryogenesis and the effects of kinetochore component modifications on proper chromosome segregation. Finally, the advantages and limitations of the CENH3-mediated haploidization approach and its applications are highlighted.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dwivedi SL et al (2015) Haploids: constraints and opportunities in plant breeding. Biotechnol Adv 33(6 Pt 1):812–829
Ren J et al (2017) Novel technologies in doubled haploid line development. Plant Biotechnol J 15(11):1361–1370
Segui-Simarro JM (2015) Editorial: Doubled haploidy in model and recalcitrant species. Front Plant Sci 6:1175
Gilles LM et al (2017) Haploid induction in plants. Curr Biol 27(20):R1095–r1097
Kalinowska K et al (2019) State-of-the-art and novel developments of in vivo haploid technologies. Theor Appl Genet 132(3):593–605
Guha S, Maheshwari SC (1964) In vitro production of embryos from anthers of datura. Nature 204(4957):497–497
Kasha KJ, Maluszynski M (2003) Production of doubled haploids in crop plants. An introduction. In: Maluszynski M, Kasha KJ, Forster BP, Szarejko I (eds) Doubled haploid production in crop plants: a manual. Springer, Dordrecht, pp 1–4
Ishii T, Karimi-Ashtiyani R, Houben A (2016) Haploidization via chromosome elimination: means and mechanisms. Annu Rev Plant Biol 67:421–438
Bashir T et al (2018) Effect of hybridization on somatic mutations and genomic rearrangements in plants. Int J Mol Sci 19(12)
Abbott R et al (2013) Hybridization and speciation. J Evol Biol 26(2):229–246
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588
Kerbs B et al (2017) The potential role of hybridization in diversification and speciation in an insular plant lineage: insights from synthetic interspecific hybrids. AoB Plants 9(5):plx043
Chen ZJ (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14(7):471–482
Kasha KJ, Kao KN (1970) High frequency haploid production in barley (Hordeum vulgare L.). Nature 225(5235):874–876
Subrahmanyam NC, Kasha KJ (1973) Selective chromosomal elimination during haploid formation in barley following interspecific hybridization. Chromosoma 42(2):111–125
Gernand D et al (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17(9):2431–2438
Devaux P (2003) The Hordeum bulbosum (L.) method. In: Maluszynski M, Kasha KJ, Forster BP, Szarejko I (eds) Doubled haploid production in crop plants: a manual. Springer, Dordrecht, pp 15–19
Ravi M, Chan SWL (2013) Centromere-mediated generation of haploid plants. In: Jiang J, Birchler JA (eds) Plant centromere biology. Wiley, Berlin, pp 169–181
Finch RA (1983) Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma 88(5):386–393
Musacchio A, Desai A (2017) A molecular view of kinetochore assembly and function. Biology 6(1)
Ingouff M et al (2010) Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20(23):2137–2143
Fu S et al (2018) Maternal doubled haploid production in interploidy hybridization between Brassica napus and Brassica allooctaploids. Planta 247(1):113–125
Gupta SB (1969) Duration of mitotic cycle and regulation of DNA replication in nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Can J Genet Cytol 11(1):133–142
Sanei M et al (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A 108(33):E498–E505
Laurie DA, Bennett MD (1989) The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome 32(6):953–961
Bennett MD, Finch RA, Barclay IR (1976) The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma 54(2):175–200
Schwarzacher-Robinson T et al (1987) Genotypic control of centromere positions of parental genomes in Hordeum × Secale hybrid metaphases. J Cell Sci 87(2):291
Ravi M, Chan SW (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464(7288):615–618
Wang G, Zhang X, Jin W (2009) An overview of plant centromeres. J Genet Genomics 36(9):529–537
Potapova T, Gorbsky GJ (2017) The consequences of chromosome segregation errors in mitosis and meiosis. Biology 6(1)
Zamariola L et al (2014) Chromosome segregation in plant meiosis. Front Plant Sci 5:279
Oliveira LC, Torres GA (2018) Plant centromeres: genetics, epigenetics and evolution. Mol Biol Rep 45(5):1491–1497
Sharma AB et al (2019) Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Res 47(3):1051–1069
Lermontova I et al (2015) Centromeric chromatin and its dynamics in plants. Plant J 83(1):4–17
Chen Y et al (2000) The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol 20(18):7037–7048
Gonzalez M et al (2014) Ectopic centromere nucleation by CENP-A in fission yeast. Genetics 198(4):1433–1446
Lermontova I et al (2011) Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J 68(1):40–50
Ravi M et al (2011) Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet 7(6):e1002121
Maheshwari S et al (2015) Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet 11(1):e1004970
Britt AB, Kuppu S (2016) Cenh3: an emerging player in haploid induction technology. Front Plant Sci 7:357
Malik HS, Henikoff S (2003) Phylogenomics of the nucleosome. Nat Struct Biol 10(11):882–891
Morey L et al (2004) The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot Cell 3(6):1533–1543
Lermontova I et al (2006) Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18(10):2443–2451
Vermaak D, Hayden HS, Henikoff S (2002) Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol 22(21):7553–7561
Black BE et al (2004) Structural determinants for generating centromeric chromatin. Nature 430(6999):578–582
Watts A, Kumar V, Bhat SR (2016) Centromeric histone H3 protein: from basic study to plant breeding applications. J Plant Biochem Biotechnol 25(4):339–348
Talbert PB et al (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14(5):1053–1066
Maehara K, Takahashi K, Saitoh S (2010) CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol 30(9):2090–2104
Raychaudhuri N et al (2012) Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol 10(12):e1001434
Caussinus E, Kanca O, Affolter M (2011) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 19(1):117–121
Heun P et al (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10(3):303–315
Hewawasam G et al (2010) Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40(3):444–454
Ranjitkar P et al (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40(3):455–464
Collins KA, Furuyama S, Biggins S (2004) Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol 14(21):1968–1972
Ravi M et al (2010) The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics 186(2):461–471
Feng C et al (2020) The deposition of CENH3 in maize is stringently regulated. Plant J 102(1):6–17
Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9(12):923–937
Stellfox ME, Bailey AO, Foltz DR (2013) Putting CENP-A in its place. Cell Mol Life Sci 70(3):387–406
Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3(4):18
Ogura Y et al (2004) Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet Syst 79(3):139–144
Kato H et al (2013) A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340(6136):1110–1113
Dawe RK et al (1999) A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11(7):1227–1238
Ishii T et al (2020) Unequal contribution of two paralogous centromeric histones to function the cowpea centromere. bioRxiv:2020.01.07.897074
Marques A et al (2016) Restructuring of holocentric centromeres during meiosis in the plant Rhynchospora pubera. Genetics 204(2):555–568
Kwon M-S et al (2007) CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell 18(6):2155–2168
Howman EV et al (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A 97(3):1148–1153
Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6(2):e1000835
Hori T et al (2017) Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Dev Cell 42(2):181–189.e3
Fujita Y et al (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12(1):17–30
Dambacher F et al (2015) Reducing proactive aggression through non-invasive brain stimulation. Soc Cogn Affect Neurosci 10(10):1303–1309
Moree B et al (2011) CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194(6):855–871
Maddox PS et al (2007) Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176(6):757–763
Lermontova I et al (2013) Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 25(9):3389–3404
Sandmann M et al (2017) Targeting of Arabidopsis KNL2 to centromeres depends on the conserved CENPC-k motif in its C terminus. Plant Cell 29(1):144–155
Kral L (2015) Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. F1000 Res 4:474–474
Dambacher S et al (2012) CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3(1):101–110
French BT, Straight AF (2019) CDK phosphorylation of Xenopus laevis M18BP1 promotes its metaphase centromere localization. EMBO J 38(4)
Batzenschlager M et al (2015) Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc Natl Acad Sci 112(28):8656–8660
Le Goff S et al (2020) The H3 histone chaperone NASPSIM3 escorts CenH3 in Arabidopsis. Plant J 101(1):71–86
Wang N, Dawe RK (2018) Centromere size and its relationship to haploid formation in plants. Mol Plant 11(3):398–406
Pickering RA (1985) Partial control of chromosome elimination by temperature in immature embryos of Hordeum vulgare L. x H. bulbosum L. Euphytica 34(3):869–874
Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140(1):136–147
Comai L (2014) Genome elimination: translating basic research into a future tool for plant breeding. PLoS Biol 12(6):e1001876
Kelliher T et al (2016) Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front Plant Sci 7:414–414
Ravi M et al (2014) A haploid genetics toolbox for Arabidopsis thaliana. Nat Commun 5:5334
Karimi-Ashtiyani R et al (2015) Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc Natl Acad Sci U S A 112(36):11211–11216
Ishii T et al (2015) The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosom Res 23(2):277–284
Van der Veken J et al (2019) Cichorium intybus L. × Cicerbita alpina Walbr.: doubled haploid chicory induction and CENH3 characterization. Euphytica 215(7):134
Kuppu S et al (2015) Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet 11(9):e1005494
Kuppu S et al (2020) A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol J 18:2068–2080
Woo JW et al (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33(11):1162–1164
Coe EH (1959) A line of maize with high haploid frequency. Am Nat 93(873):381–382
Prigge V et al (2012) New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 190(2):781–793
Wang S, Jin W, Wang K (2019) Centromere histone H3- and phospholipase-mediated haploid induction in plants. Plant Methods 15(1):42
Kelliher T et al (2017) MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542(7639):105–109
Gilles LM et al (2017) Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J 36(6):707–717
Liu C et al (2017) A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in Maize. Mol Plant 10(3):520–522
Yao L et al (2018) OsMATL mutation induces haploid seed formation in indica rice. Nat Plants 4(8):530–533
Zhong Y et al (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat Plants 5(6):575–580
Khanday I et al (2019) A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565(7737):91–95
Ron M et al (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166(2):455–469
Ron M, Kuppu S, Britt AB (2019) Cenh3 deletion mutants. Patent WO 2019/136417A2
Camp RHMOD et al (2017) Method for the production of haploid and subsequent doubled haploid plants field of the invention. Patent WO 2017/058022A1
Van Dun CMP, Lelivelt CLC, Movahedi S (2017) Non-transgenic haploid inducer lines in cucurbits. Patent WO 2017/081011
Dirks R et al (2009) Reverse breeding: a novel breeding approach based on engineered meiosis. Plant Biotechnol J 7(9):837–845
Wijnker E et al (2014) Hybrid recreation by reverse breeding in Arabidopsis thaliana. Nat Protoc 9(4):761–772
Spillane C, Curtis MD, Grossniklaus U (2004) Apomixis technology development-virgin births in farmers’ fields? Nat Biotechnol 22(6):687–691
Mieulet D et al (2016) Turning rice meiosis into mitosis. Cell Res 26(11):1242–1254
d’Erfurth I et al (2009) Turning meiosis into mitosis. PLoS Biol 7(6):e1000124
Marimuthu MP et al (2011) Synthetic clonal reproduction through seeds. Science 331(6019):876
Kelliher T et al (2019) One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37(3):287–292
Wang B et al (2019) Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol Plant 12(4):597–602
Cortes-Silva N et al (2020) CenH3-Independent kinetochore assembly in Lepidoptera requires CCAN, including CENP-T. Curr Biol 30(4):561–572.e10
Drinnenberg IA et al (2014) Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. elife 3
Acknowledgments
The author is very grateful to Dr. Ali Mohammad Banaei-Moghaddam, Institute of Biochemistry and Biophysics (IBB), University of Tehran, for valuable suggestions and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Karimi-Ashtiyani, R. (2021). Centromere Engineering as an Emerging Tool for Haploid Plant Production: Advances and Challenges. In: Segui-Simarro, J.M. (eds) Doubled Haploid Technology. Methods in Molecular Biology, vol 2289. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1331-3_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1331-3_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1330-6
Online ISBN: 978-1-0716-1331-3
eBook Packages: Springer Protocols