Abstract
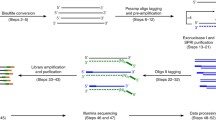
Single-cell bisulfite sequencing (scBS-seq) enables profiling of DNA methylation at single-nucleotide resolution and across all genomic features. It can explore methylation differences between cells in mixed cell populations and profile methylation in very rare cell types, such as mammalian oocytes and cells from early embryos. Here, we outline the scBS-seq protocol in a 96-well plate format applicable to studies of moderate throughput.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W (2016) Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol 17:72. https://doi.org/10.1186/s13059-016-0944-x
Kelsey G, Stegle O, Reik W (2017) Single-cell epigenomics: recording the past and predicting the future. Science 358(6359):69–75. https://doi.org/10.1126/science.aan6826
Guo H, Zhu P, Wu X, Li X, Wen L, Tang F (2013) Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res 23(12):2126–2135. https://doi.org/10.1101/gr.161679.113
Guo H, Zhu P, Yan L et al (2014) The DNA methylation landscape of human early embryos. Nature 511(7511):606–610. https://doi.org/10.1038/nature13544
Miura F, Enomoto Y, Dairiki R, Ito T (2012) Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40(17):e136. https://doi.org/10.1093/nar/gks454
Smallwood SA, Lee HJ, Angermueller C et al (2014) Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 11(8):817–820. https://doi.org/10.1038/nmeth.3035
Nashun B, Hill PWS, Smallwood SA et al (2015) Hira is essential for normal transcriptional regulation and efficient de novo DNA methylation during mouse oogenesis. Mol Cell 60(4):611–625. https://doi.org/10.1016/j.molcel.2015.10.010
Zhu P, Guo H, Ren Y et al (2018) Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet 50(X):12–19. https://doi.org/10.1038/s41588-017-0007-6
Farlik M, Sheffield NC, Nuzzo A et al (2015) Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep 10(8):1386–1397. https://doi.org/10.1016/j.celrep.2015.02.001
Gravina S, Dong X, Yu B, Vijg J (2016) Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome Biol 17(1):150. https://doi.org/10.1186/s13059-016-1011-3
Clark SJ, Smallwood SA, Lee HJ, Krueger F, Reik W, Kelsey G (2017) scBS-seq: genome-wide base-resolution mapping of DNA methylation in single cells. Nat Protoc 12(3):534–547. https://doi.org/10.1038/nprot.2016.187
Mulqueen RM, Pokholok D, Norberg SJ et al (2018) Highly scalable generation of DNA methylation profiles in single cells. Nat Biotechnol 36(5):428–431. https://doi.org/10.1038/nbt.4112
Hu Y, Huang K, An Q et al (2016) Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol 7:88. https://doi.org/10.1186/s13059-016-0950-z
Hu Y, Guo H, Cao C et al (2016) Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26(3):304–319. https://doi.org/10.1038/cr.2016.23
Angermueller C, Clark SJ, Lee HJ et al (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13(3):229–232. https://doi.org/10.1038/nmeth.3728
Guo F, Li L, Li J et al (2017) Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res 27(8):967–988. https://doi.org/10.1038/cr.2017.82
Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA (2012) Genome-wide mapping of nucleosome positioning and DNA methylation within individual molecules. Genome Res 22(12):2497–2506. https://doi.org/10.1101/gr.143008
Clark SJ, Argelaguet R, Kapourani C-A et al (2018) Joint profiling of chromatin accessibility, DNA methylation and transcription in single cells. Nat Commun 9(1):781. https://doi.org/10.1038/s41467-018-03149-4
Gu C, Liu S, Wu Q, Zhang L, Guo F (2019) Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res 29(2):110–123. https://doi.org/10.1038/s41422-018-0125-4
Li L, Guo F, Gao Y et al (2018) Single-cell multi-omics sequencing of early human embryos. Nat Cell Biol 20(7):847–858. https://doi.org/10.1038/s41556-018-0123-2
Argelaguet R, Mohammed H, Clark SJ et al (2019) Single cell multi-omics profiling reveals a hierarchical epigenetic landscape during mammalian germ layer specification. Nature 576(7787):487–491. https://doi.org/10.1038/s41586-019-1825-8
Lee HJ, Smallwood SA (2018) Genome-wide analysis of DNA methylation in single cells using a post-bisulfite adapter tagging approach. Methods Mol Biol 1712:87–95. https://doi.org/10.1007/978-1-4939-7514-3_7
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Galvão, A., Kelsey, G. (2021). Profiling DNA Methylation Genome-Wide in Single Cells. In: Ancelin, K., Borensztein, M. (eds) Epigenetic Reprogramming During Mouse Embryogenesis. Methods in Molecular Biology, vol 2214. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0958-3_15
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0958-3_15
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0957-6
Online ISBN: 978-1-0716-0958-3
eBook Packages: Springer Protocols