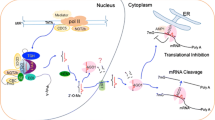
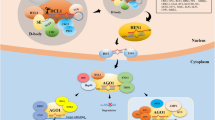
Abstract
MicroRNAs control plant development and are key regulators of plant responses to biotic and abiotic stresses. Thus, their expression must be carefully controlled since both excess and deficiency of a given microRNA may be deleterious to plant cell. MicroRNA expression regulation can occur at several stages of their biogenesis pathway. One of the most important of these regulatory checkpoints is transcription efficiency. mirEX database is a tool for exploration and visualization of plant pri-miRNA expression profiles. It includes results obtained using high-throughput RT-qPCR platform designed to monitor pri-miRNA expression in different miRNA biogenesis mutants and developmental stages of Arabidopsis, barley, and Pellia plants. A step-by-step instruction for browsing the database and detailed protocol for high-throughput RT-qPCR experiments, including list of primers designed for the amplification of pri-miRNAs, are presented.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44. https://doi.org/10.1146/annurev.cellbio.042308.113417
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203. https://doi.org/10.1016/j.tplants.2012.01.010
Barciszewska-Pacak M, Milanowska K, Knop K, Bielewicz D, Nuc P, Plewka P, Pacak AM, Vazquez F, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z (2015) Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front Plant Sci 6:410. https://doi.org/10.3389/fpls.2015.00410
Smoczynska A, Szweykowska-Kulinska Z (2017) MicroRNA-mediated regulation of flower development in grasses. Acta Biochim Pol 63:687–692. https://doi.org/10.18388/abp.2016_1358
Yu Y, Jia T, Chen X (2017) The ‘how’ and ‘where’ of plant microRNAs. New Phytol 216:1002–1017. https://doi.org/10.1111/nph.14834
Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138:2145–2154. https://doi.org/10.1104/pp.105.062943
Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495. https://doi.org/10.1016/S0960-9822(02)01017-5
Stepien A, Knop K, Dolata J, Taube M, Bajczyk M, Barciszewska-Pacak M, Pacak A, Jarmolowski A, Szweykowska-Kulinska Z (2017) Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip Rev RNA 8(3). https://doi.org/10.1002/wrna.1403
Zielezinski A, Dolata J, Alaba S, Kruszka K, Pacak A, Swida-Barteczka A, Knop K, Stepien A, Bielewicz D, Pietrykowska H (2015) mirEX 2.0-an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol 15:144. https://doi.org/10.1186/s12870-015-0533-2
Lepe-Soltero D, Armenta-Medina A, Xiang D, Datla R, Gillmor CS, Abreu-Goodger C (2017) Annotating and quantifying pri-miRNA transcripts using RNA-Seq data of wild type and serrate-1 globular stage embryos of Arabidopsis thaliana. Data Brief 15:642–647. https://doi.org/10.1016/j.dib.2017.10.019
Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC (2012) Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 48:521–531. https://doi.org/10.1016/j.molcel.2012.08.032
Bielewicz D, Kalak M, Kalyna M, Windels D, Barta A, Vazquez F, Szweykowska-Kulinska Z, Jarmolowski A (2013) Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep 14:622–628. https://doi.org/10.1038/embor.2013.62
Schwab R, Speth C, Laubinger S, Voinnet O (2013) Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep 14:615–621. https://doi.org/10.1038/embor.2013.58
Barciszewska-Pacak M, Knop K, Jarmołowski A, Szweykowska-Kulińska Z (2016) Arabidopsis thaliana microRNA162 level is posttranscriptionally regulated via splicing and polyadenylation site selection. Acta Biochim Pol 63:811–816. https://doi.org/10.18388/abp.2016_1349
Knop K, Stepien A, Barciszewska-Pacak M, Taube M, Bielewicz D, Michalak M, Borst JW, Jarmolowski A, Szweykowska-Kulinska Z (2017) Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res 45:2757–2775. https://doi.org/10.1093/nar/gkw895
Szweykowska-Kulinska Z, Jarmolowski A, Vazquez F (2013) The crosstalk between plant microRNA biogenesis factors and the spliceosome. Plant Signal Behav 8(11):e26955. https://doi.org/10.4161/psb.26955
Kruszka K, Pacak A, Swida-Barteczka A, Stefaniak AK, Kaja E, Sierocka I, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z (2013) Developmentally regulated expression and complex processing of barley pri-microRNAs. BMC Genomics 14:34. https://doi.org/10.1186/1471-2164-14-34
Trevino V, Falciani F, Barrera-Saldana HA (2007) DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol Med 13:527–541. https://doi.org/10.2119/2006-00107.Trevino
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517. https://doi.org/10.1101/gr.079558.108
Everaert C, Luypaert M, Maag JL, Cheng QX, Dinger ME, Hellemans J, Mestdagh P (2017) Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep 7:1559. https://doi.org/10.1038/s41598-017-01617-3
Prigge MJ, Wagner DR (2001) The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13:1263–1279. https://doi.org/10.1105/TPC.010095
Weaver S, Dube S, Mir A, Qin J, Sun G, Ramakrishnan R, Jones RC, Livak KJ (2010) Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 50:271–276. https://doi.org/10.1016/j.ymeth.2010.01.003
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17. https://doi.org/10.1104/pp.105.063743
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618. https://doi.org/10.1111/j.1467-7652.2008.00346.x
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20:1734–1735. https://doi.org/10.1105/tpc.108.059774
Rapacz M, Stępień A, Skorupa K (2012) Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol Plant 34:1723–1733
Bielewicz D, Dolata J, Zielezinski A, Alaba S, Szarzynska B, Szczesniak MW, Jarmolowski A, Szweykowska-Kulinska Z, Karlowski WM (2012) mirEX: a platform for comparative exploration of plant pri-miRNA expression data. Nucleic Acids Res 40:D191–D197. https://doi.org/10.1093/nar/gkr878
Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130:808–822. https://doi.org/10.1104/pp.003491
Devaux P, Adamski P, Surma M (1992) Inheritance of seed set in crosses of spring barley and Hordeum bulbosum L. Crop Sci 32:269–271. https://doi.org/10.2135/cropsci1992.0011183X003200010054x
Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage–based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13:1499–1510. https://doi.org/10.1105/TPC.010011
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421. https://doi.org/10.1111/j.1365-3180.1974.tb01084.x
Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. https://doi.org/10.1093/nar/gkp045
Dolata J, Bajczyk M, Bielewicz D, Niedojadlo K, Niedojadlo J, Pietrykowska H, Walczak W, Szweykowska-Kulinska Z, Jarmolowski A (2016) Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol 172:297–312. https://doi.org/10.1104/pp.16.00830
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(− ΔΔCT) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Acknowledgements
The project is supported by the National Science Centre, Poland (NCN): UMO-2017/25/B/NZ1/00603, UMO-2016/23/D/NZ1/00152, UMO-2016/21/B/NZ9/00550, UMO-2016/23/B/NZ9/00857, 2013/10/A/NZ1/00557, UMO-2016/23/B/NZ9/00862, by the Foundation for Polish Science (grant START 2017 to Agata S) and by KNOW RNA Research Centre in Poznań (No. 01/KNOW2/2014).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Dolata, J. et al. (2021). Quantitative Analysis of Plant miRNA Primary Transcripts. In: Jin, H., Kaloshian, I. (eds) RNA Abundance Analysis . Methods in Molecular Biology, vol 2170. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0743-5_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0743-5_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0742-8
Online ISBN: 978-1-0716-0743-5
eBook Packages: Springer Protocols