Abstract
The selective immobilization of proteins represents an essential step in the selection of binding proteins such as antibodies. The immobilization strategy determines how the target protein is presented to the binders and thereby directly affects the experimental outcome. This poses specific challenges for membrane proteins due to their inherent lack of stability and limited exposed hydrophilic surfaces. Here we detail methodologies for the selective immobilization of membrane proteins based on the strong biotin-avidin interaction and with a specific focus on its application for the selection of nanobodies and sybodies. We discuss the challenges in generating and benefits of obtaining an equimolar biotin to target-protein ratio.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Biotin
- Neutravidin
- Streptavidin
- FX cloning
- Avi-tag
- Panning
- Phage display
- Ribosome display
- Nanobodies
- Sybodies
1 Introduction
Antibody fragments and in particular nanobodies have become indispensable tools for studying structural and functional aspects of membrane proteins [1]. The generation of these binders involves the stringent phenotypic selection of individual members from libraries holding many variants. Central to this procedure is the selective immobilization of the target protein to enrich those members of the library that specifically interact with it. We recently developed an in vitro selection platform based on three large synthetic nanobody (sybody) libraries that allows the generation of binders under entirely defined and mild conditions compatible with membrane proteins [2]. A major hallmark of our platform is its optimization toward the routine selection of binders against membrane proteins, which entails successive alterations in display technology, immobilization surface, and the application of solution panning. The latter allows the free target protein to interact with the displayed binders in solution, preceding a rapid (within minutes) immobilization on beads and subsequent pull-down of the target protein-binder complexes. Hereby delicate membrane proteins are protected from denaturation resulting from prolonged exposure to surfaces at high protein densities. Hence, the selective immobilization of the target protein is a key step in selection procedures.
Though seemingly trivial, the choice of the immobilization strategy is of great relevance as this may dramatically skew the selection and directly affect the quality and quantity of unique binders identified. Given the aim of obtaining multiple strong binders against different, three-dimensional epitopes, an ideal protein immobilization strategy should: (1) preserve the native three-dimensional structure; (2) allow a non-oriented, ideally random orientation of the target protein with high accessibility of potential epitopes; (3) capture the target protein selectively, rapidly (within a few minutes), and stably (over prolonged periods of several hours) in a variety of buffer conditions and a broad temperature range; and (4) allow near-complete capture of the target protein to avoid loss of binder diversity during solution panning. In addition, the strategy should not interfere with biogenesis and function of the target protein and should be facile to implement. Among the multitude of protein immobilization strategies [3], the biotin/avidin-based interaction fits these criteria best and is therefore widely used [4].
The interaction between the vitamin biotin and avidin or its variants streptavidin and neutravidin is one of the strongest non-covalent interactions known (Kd of ~10−14 M) and has a half-life of several days [5, 6]. The interaction remains stable over a broad range of temperatures [7], pH values, and denaturants [8, 9]. Avidin, streptavidin, and neutravidin are homotetrameric proteins with four biotin-binding sites. Streptavidin, derived from bacterial origin, and neutravidin, a deglycosylated form of avidin, are generally preferred over avidin, as the absence of glycosylation and their lower pI values reduce nonspecific binding [5, 8]. Importantly, naturally biotinylated proteins are rare: in E. coli or mammalian cells the number of proteins holding a covalently attached biotin amount to one and four, respectively [10, 11].
Biotinylation of a target membrane protein can be achieved chemically or enzymatically. Chemical biotinylation is most conveniently done by targeting the primary amine of a surface-exposed lysine residue using biotin derivatized with an N-hydroxysuccinimide (NHS) group. This reaction can be performed under comparably mild, biocompatible conditions. Due to the general abundance of lysines on protein surfaces, amine chemistry allows the introduction of biotin at different positions in the protein. Consequently, the target protein can be immobilized in several orientations allowing exposure of different potential epitopes, provided that only one biotin group is introduced. A higher degree of labeling is disadvantageous as this may restrict flexibility and surface presentation and may even directly interfere with binding of the antibody by masking the epitope. As an alternative to the comparably abundant lysines, cysteines may be targeted using, e.g., biotin derivatized with a maleimide group. The main advantage of chemical biotinylation is the random target orientation during immobilization. This comes at the price of two disadvantages: chemical biotinylation typically results in a distribution of target proteins carrying none, one, or multiple biotin moieties; and biotinylation of lysines may modify, and thereby mask, potential epitopes.
The E. coli biotin protein ligase BirA requires biotin and ATP to biotinylate its only target, the biotin carboxyl carrier protein (BCCP) subunit of acetyl-CoA carboxylase, at a specific lysine in an evolutionary conserved amino acid sequence. Engineering of this sequence led to the identification of the Avi-tag, a 15 amino acid stretch, GLNDIFEAQ-K-IEWHE, that is biotinylated with high efficiency [12, 13]. Avi-tags fused to the N- or C-terminus [14] or even integrated in exposed loops [15] are efficiently biotinylated by BirA. Enzymatic biotinylation of membrane proteins can be done in vivo using native or co-expressed BirA or in vitro using the purified BirA protein. The main advantage of enzymatic biotinylation is its high efficiency and specificity, resulting in nearly complete and exclusive biotinylation of the lysin residue in the Avi-tag. Hence, the highly desirable biotin to target protein ratio of 1:1 can easily be achieved. However, enzymatic biotinylation has two major disadvantages: all target proteins are immobilized in the same orientation, which may render some epitopes inaccessible; this problem is exacerbated for homo-oligomeric target proteins, where several biotin moieties are introduced via the Avi-tag; and the attachment of the Avi-tag sequence to the open reading frame of the target protein requires molecular cloning and potentially construct optimization.
The biochemical quality of the membrane protein target is arguably the most critical parameter when performing binder selections. Hence, it is paramount that the biotinylation procedure does not compromise the structure and function of the target protein. Therefore, biotinylated target proteins need to be experimentally tested for activity and structural integrity using size exclusion chromatography, both for enzymatic and chemical biotinylation.
This chapter first details a facile cloning strategy for fusing sequences for N- or C-terminal Avi-tags to the target open reading frame. Subsequently, we describe approaches for enzymatic and chemical biotinylation of (Avi-tagged) membrane proteins and conclude with methodology to assess the degree of biotinylation. Together, this chapter provides all relevant information required to selectively immobilize membrane proteins using the biotin/avidin interaction.
2 Materials
2.1 FX Cloning
-
1.
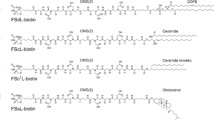
FX cloning vectors. E. coli expression vectors for the arabinose-controlled PBAD promoter [16] and holding sequences coding for an Avi-tag in combination with an HRV 3C protease cleavable GFP-His-tag or His-tag (Fig. 1) are available on Addgene (#47069, #47071-47075). The optional intermediate vector pINIT_cat for subcloning is available on Addgene as well (#46858). All FX cloning vectors should be propagated in a CcdB-resistant E. coli strain such as DB3.1 [17].
-
2.
Dedicated forward and reverse primers targeting the gene of interest and compatible with FX cloning. For ordering, choose the smallest synthesis scale and mere desalting as purification.
-
3.
Template DNA for the open reading frame of interest (e.g., genomic DNA or plasmid).
-
4.
Phusion DNA polymerase, corresponding buffer, and dNTPs.
-
5.
TAE buffer, TAE agarose gel, and agarose gel DNA extraction kit.
-
6.
SapI restriction enzyme and corresponding buffer.
-
7.
T4 DNA ligase.
-
8.
10 mM ATP: 10 mM Na2-ATP, 10 mM MgSO4. Dissolve in 50 mM KPi, pH 7.0 and adjust to pH 6.5–7.0 with NaOH. Store in small aliquots at −20 °C.
-
9.
CcdB-sensitive E. coli strain (e.g., MC1061 [18]).
-
10.
LB medium and LB-agar plate supplemented with the appropriate antibiotic. For ampicillin (Amp) and chloramphenicol (Cam), use 100 μg/mL and 34 μg/mL, respectively.
-
11.
Miniprep kit (QIAGEN).
-
12.
Sequencing primers for pINIT_cat or the arabinose-controlled PBAD expression vectors (Table 1).
Protein biotinylation toolkit. (a) FX cloning expression vectors for the introduction of Avi-tags. (b) Assessing the degree of biotinylation by a streptavidin-induced mobility shift in SDS-PAGE. Avi-tagged OmpA of Klebsiella pneumoniae and MBP of E. coli were enzymatically biotinylated in vitro using purified BirA. (c) GFP (126 μM) was chemically biotinylated with a fivefold molar excess of EZ-Link Sulfo-NHS-LC-Biotin (630 μM) in PBS for 30 min at 25 °C. The resulting biotinylation pattern was determined by mass spectrometry (ESI-MS). (d) Quantification of the biotinylation pattern. Less than 5% of GFP was devoid of biotin. The data were fitted with a Gaussian curve
2.2 BirA-Based In Vitro Biotinylation
-
1.
BirA at 8 mg/mL (228 μM) in 50 mM Tris–HCl pH 7.5, 200 mM KCl, 50% glycerol, 0.1 mM DTT. His-tagged BirA can be produced using pET21a-BirA (Addgene) as described [19] but it is also commercially available (e.g., from Sigma-Aldrich). Store for prolonged periods at −80 °C. Substocks stored at −20 °C will remain liquid and ready to use.
-
2.
HRV 3C protease at 6 mg/mL (276 μM) in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM TCEP, and 50% glycerol. His-tagged 3C protease can be produced as described [20] but is also commercially available (e.g., from Sigma-Aldrich). Store 100 μL aliquots at −80 °C.
-
3.
200 mM ATP (dissolve in 50 mM KPi, pH 7.0 and adjust to pH 6.5–7.0 with NaOH).
-
4.
1 mM biotin in 50 mM Bicine buffer pH 8.3. Store in small aliquots at −20 °C.
-
5.
1 M MgOAc.
-
6.
Ni-NTA resin or prepacked Ni-NTA column.
2.3 BirA-Based In Vivo Biotinylation
-
1.
Expression vectors. Mammalian vectors for protein expression based on a pCDNA3.1(+) backbone (Thermo Fisher) controlled by a CMV promoter and holding a Kozak sequence preceding sequence coding for an N- (pC039) or C-terminal (pC031) Avi-tag in combination with an HRV 3C protease cleavable GFP-His-tag, respectively (Fig. 1).
-
2.
BirA co-expression vectors. Co-expression of E. coli BirA in mammalian cell culture and under control of a CMV promoter is assured by the presence of an additional plasmid coding for BirA with a C-terminal Myc-tag for intracellular biotinylation, or BirA with an N-terminal IgH signal sequence and C-terminal KDEL ER-retention signal for biotinylation of extracellularly located Avi-tags.
-
3.
Opti-MEM reduced serum medium (Thermo Fisher).
-
4.
ExpiFectamine 293 transfection kit (Thermo Fisher; part of the expression kit).
-
5.
Expi293 expression medium (Thermo Fisher) and Freestyle medium (Thermo Fisher).
-
6.
Fernbach cell culture shaking flask (e.g., 3 L) plus CO2-gassed shaker platform.
-
7.
Optional: 2.1 mM biotin in Expi293 medium.
-
8.
Steritop filter (250 mL, Millipore).
2.4 Chemical Biotinylation
-
1.
PBS buffer pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4.
-
2.
EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher).
-
3.
Dimethyl sulfoxide (DMSO).
2.5 Assessing Degree of Biotinylation
-
1.
Streptavidin at 1 mg/mL in MilliQ. Store in aliquots at −20 °C.
-
2.
5× SDS-PAGE sample buffer.
-
3.
SDS-PAGE gel and setup.
3 Methods
3.1 FX Cloning
-
1.
Design FX-cloning compatible primers targeting your gene of interest. This is most conveniently done online at the https://www.fxcloning.org website using a FASTA-formatted sequence including a start and stop codon. Order the primer set optimized toward removal of stable hairpin structures (see Note 1).
-
2.
Amplify the gene of interest by PCR. Prepare a 50 μL PCR reaction and add the DNA polymerase immediately prior to starting the reaction. Use a touch-down [21] program, e.g., (1) 30 s at 98 °C; (2) 10 s at 98 °C; (3) 15 s at 61 °C (annealing temperature decreased by 0.5 °C per cycle); (4) 15–30 s/kb at 72 °C; repeat (2)–(4) 14 times; (5) 10 s at 98 °C; (6) 15 s at 53 °C; (7) 15–30 s/kb at 72 °C; repeat (5)–(7) 14 times; (8) 120 s at 72 °C; (9) unlimited at 10 °C.
-
3.
Analyze the product on a preparative TAE agarose gel. Purify the relevant band using a DNA gel extraction kit and quantify the DNA spectrophotometrically.
-
4.
Mix 50 ng of pINIT_cat with the extracted insert in a molar ratio of vector:insert of 1:5 (see Note 2).
-
5.
Add 1 μL 10× SapI buffer and adjust the volume to 9 μL with ultrapure water. Add 1 μL SapI (2 U) and incubate for 1 h at 37 °C in a PCR machine.
-
6.
Heat inactivate the SapI for 20 min at 65 °C. Let the sample cool down and add 1.25 μL 10 mM ATP and 1.25 μL T4 DNA ligase (1.25 U). Incubate for 1 h at room temperature.
-
7.
Heat inactivate the T4 DNA ligase for 20 min at 65 °C and transform 5 μL of the ligation mix to 100 μL chemically competent E. coli MC1061 cells (or an alternative CcdB-sensitive strain).
-
8.
Plate 10% and 90% aliquots on LB-agar-Cam plates and incubate overnight at 37 °C.
-
9.
Use a single colony to inoculate 5 mL LB-Cam and cultivate overnight. Isolate the plasmid using a miniprep kit, determine the concentration spectrophotometrically, and verify the insert by DNA sequencing using the pINIT_cat sequencing primers.
-
10.
Mix 50 ng of the FX cloning Avi-tag expression vector (see Note 3) with pINIT_cat carrying the insert to a molar ratio of vector:pINIT_cat-derivative of 1:5. Add 1 μL 10× SapI buffer and adjust the volume to 9 μL with ultrapure water. Add 1 μL SapI (2 U) and incubate for 1 h at 37 °C in a PCR machine.
-
11.
Heat inactivate the SapI for 20 min at 65 °C. Let the sample cool down and add 1.25 μL 10 mM ATP and 1.25 μL T4 DNA ligase (1.25 U). Incubate for 1 h at room temperature.
-
12.
Heat inactivate the T4 DNA ligase for 20 min at 65 °C and transform 5 μL of the ligation mix to 100 μL chemically competent E. coli MC1061 cells.
-
13.
Plate 10% and 90% aliquots on LB-agar-Amp plates. Incubate the plates overnight at 37 °C.
-
14.
Use a single colony to inoculate 5 mL LB-Amp and cultivate overnight at 37 °C.
-
15.
Archive the culture as a glycerol stock at −80 °C (see Note 4). This stock can serve for inoculation of expression cultures based on the araBAD promoter (see Notes 5 and 6).
3.2 BirA-Based In Vitro Biotinylation
-
1.
Recombinantly express the target protein using previously established procedures [22] (see Note 7). Purify the Avi-tagged target protein (see Note 8) and determine the protein concentration spectrophotometrically.
-
2.
Add 3C protease to a molar ratio of 1:10 to cleave off the decaHis-tag while dialyzing the sample for 1 h at 4 °C to remove excess imidazole (see Note 9).
-
3.
Adjust the target protein concentration to 10–50 μM (either by dilution or using a concentrator unit). Add biotin to a molar ratio of target protein:biotin of 1:1.5, 5 mM ATP, 10 mM MgOAc and BirA to a molar ratio of target protein:BirA of 20:1 (see Note 10). Incubate the sample overnight at 4 °C (see Note 11).
-
4.
Remove His-tagged BirA, HRV 3C protease, and potential remaining contaminants from the sample by reverse IMAC and collect the flow-through holding the biotinylated target protein.
-
5.
Perform size exclusion chromatography (SEC) to remove soluble aggregates and excess of biotin from the sample (see Note 12). Determine the degree of biotinylation as outlined in Subheading 3.5.
-
6.
Proceed with the selection of binders such as nanobodies and sybodies (see Note 13) or store the target protein (see Notes 14 and 15).
3.3 BirA-Based In Vivo Biotinylation
-
1.
Generate mammalian expression vectors for the gene of interest in pC031 or pC039 to obtain a fusion protein with an N- or C-terminal Avi-tag (see Notes 16 and 17).
-
2.
Split an Expi293 subculture (see Note 18), typically grown to 3-5 × 106 cells/mL, into a 3 L Fernbach shaking flask and adjust to a final volume of 0.6 L Expi293 medium with a density of 0.7 × 106 cells/mL.
-
3.
Incubate the culture for 72 h at 37 °C, under humidified atmosphere and 5% CO2 in a shaking incubator.
-
4.
On the day of the transient transfection, adjust the culture to 830 mL with a density of 3.4 × 106 cells/mL by adding Expi293 medium and/or removing cells.
-
5.
Add 20 mL biotin solution (final concentration of 50 μM) (see Note 19).
-
6.
Pipet 50 mL Opti-MEM into a 100 mL sterile Schott bottle. Add 2.7 mL ExpiFectamine transfection reagent, shake gently, and incubate for 5 min at room temperature.
-
7.
Pipet 50 mL Opti-MEM into a second 100 mL Schott bottle and add the two plasmid batches in a final amount of 1 mg to 0.1 mg, target protein expression plasmid:BirA expression plasmid, respectively. Shake gently and incubate for 5 min at room temperature.
-
8.
Mix the contents of both bottles, filter sterilize, and incubate for 20–30 min at room temperature to form the transfection complex.
-
9.
Add 100 mL of the formed transfection complex to the Fernbach shaking flask with 850 mL of cell culture for a final volume of 950 mL. Incubate further at 37 °C and 5% CO2 with mild shaking.
-
10.
Add sterile 5 mL Enhancer 1 and 50 mL Enhancer 2 from the ExpiFectamine 293 transfection kit at 16–20 h post-transfection and continue incubation.
-
11.
Incubate for a total time of approximately 48-72 h post-transfection depending on the most optimal condition for protein expression. Harvest the cells by centrifugation at 3000 × g for 15 min, flash freeze the pellet in liquid nitrogen, and store at −80 °C.
-
12.
Purify the biotinylated Avi-tagged target protein (see Note 8) and determine the protein concentration spectrophotometrically. Determine the degree of biotinylation as outlined in Subheading 3.5. Proceed with the selection of binders such as nanobodies and sybodies (see Note 13) or store the target protein (see Notes 14 and 15).
3.4 Chemical Biotinylation
-
1.
Recombinantly express the target protein using previously established procedures [22]. Purify the target protein and employ preparative SEC using PBS, supplemented with the required detergent, as buffer (see Note 20). Determine the protein concentration spectrophotometrically.
-
2.
Concentrate the target protein to 50-200 μM.
-
3.
Dissolve EZ-Link Sulfo-NHS-LC-Biotin in highly pure DMSO to a concentration of 10 mM (see Note 21).
-
4.
Add EZ-Link Sulfo-NHS-LC-Biotin to the target protein in fivefold molar excess and incubate at 25 °C for 30 min under gentle agitation (see Note 22).
-
5.
Perform SEC to remove excess of biotin from the sample (see Note 12).
-
6.
Determine the biotinylation pattern of the biotinylated target protein by mass-spectrometry (Fig. 1, see Note 23). In case mass spectrometry analysis is not available or cannot be carried out due to the target’s high molecular weight, determine the degree of biotinylation as outlined in Subheading 3.5.
-
7.
Proceed with the selection of binders such as nanobodies and sybodies (see Note 13) or store the target protein (see Notes 14 and 15).
3.5 Assessing Degree of Biotinylation
-
1.
Mix two aliquots of 10 μg biotinylated target protein with 5× SDS-PAGE sample buffer.
-
2.
Add streptavidin to one of the aliquots in a 1:1 molar ratio of target protein:streptavidin (see Note 24).
-
3.
Analyze the control (no addition) and test (streptavidin addition) samples in adjacent lanes on SDS-PAGE.
-
4.
Stain the gel with Coomassie Brilliant Blue R-250 and quantify the band intensities with the ImageJ software and calculate the degree of biotinylation (see Notes 25 and 26).
4 Notes
-
1.
As the N- and C-termini of most proteins are comparably long and flexible, we generally do not insert a linker sequence between the target protein and the Avi-tag. Nevertheless, should this be desired, a sequence for a linker is best introduced at this step.
-
2.
Alternatively, should subcloning of a sequence-verified open reading frame not be required, proceed with Step 10 and use the purified PCR product to replace the pINIT_cat holding the insert.
-
3.
Should an expression and purification strategy for the protein already be established, this combination of tags and fusions proteins should guide the choice for the expression vector. We recommend the production of protein variants with N- and C-terminal Avi-tags as this may allow the presentation of different surfaces of the target protein.
-
4.
For other expression systems that require fresh transformations for expression cultures, e.g., those based on the T7 promoter, the stock serves as a plasmid source. No additional verification by DNA sequencing is required following the subcloning of a sequence-verified open reading frame from pINIT_cat to an FX cloning expression vector.
-
5.
The FX-cloning expression vectors for fused Avi-tag allow recombinant expression in E. coli under the control of the PBAD promotor with decaHis-tag. Instead of subcloning a sequence-verified ORF from pINITIAL to an FX cloning Avi-tag expression vector, PCR products can also be cloned immediately into an FX cloning Avi-tag expression vector. This requires sequence verification of each expression vector. If multiple expression vectors are constructed, subcloning from pINITIAL prevents excessive sequencing. If one aims for only a single Avi-tagged construct, we recommend starting with pBXNH3CA (Addgene #47069), which adds a cleavable N-terminal decaHis-tag and a C-terminal Avi-tag to the protein. In our hands, this vector resulted in good expression levels for a number of ABC transporters as well as maltose-binding protein (MBP) and GFP.
-
6.
Should expression in alternative pro- or eukaryotic systems be preferred, the PBAD-based expression vectors may serve as facile intermediates for fusing the Avi-tag sequences.
-
7.
If the Avi-tagged target protein is expressed in E. coli and if the Avi-tag sequence is located in the cytoplasm, the Avi-tag will be biotinylated in vivo by virtue of the natively expressed BirA. The degree of biotinylation varies from case to case (subject to availability of biotin, level of target protein overexpression and accessibility of Avi-tag), but is often incomplete. The degree of in vivo biotinylation may be increased by co-expression of BirA and supplementation of the medium with biotin [14]. However, due to the relevance of complete biotinylation of the Avi-tag, our protocol ignores in vivo biotinylation of the target within E. coli and assures full biotinylation by performing an additional in vitro step. If required, the degree of native biotinylation can be assessed as outlined in Subheading 3.5.
-
8.
The BirA-based biotinylation protocol describes the procedure for His-tagged target protein but can in principle be adapted to protocols involving other affinity-tags. The use of strep-tags [23] or fusions with streptavidin-binding-protein (SBP) [24] should be avoided as biotinylation of Avi-tags by endogenous BirA, which may reach a very high degree depending on the experimental conditions and target protein, will prevent elution from the respective columns.
-
9.
Although BirA is inhibited by NaCl (over 100 mM) and glycerol (over 1%) [14], we generally use buffers containing 150 mM NaCl and 10% glycerol if the target membrane protein requires this for maintaining a well-folded state. We compensate for the reduced BirA activity by biotinylating for prolonged periods (overnight).
-
10.
Addition of extra amount of detergent might be necessary to keep the detergent concentration well above the CMC.
-
11.
The BirA-based biotinylation reaction can also be performed for 1 h at room temperature if the target protein is stable under these conditions. For most membrane proteins we recommend keeping the sample at 4 °C.
-
12.
Removal of free biotin is often crucial for downstream processes. In case no size exclusion chromatography is performed, use dialysis or a desalting column to remove the excess biotin from the sample.
-
13.
The outcome of the binder selection depends to a very large extent on the quality of the target protein used. Productive binder selections are expected if: (1) the SEC profile of the biotinylated target protein is monodisperse and very similar to that obtained for non-biotinylated target protein; (2) the fraction of non-biotinylated target protein is less than 10%; and (3) in case of chemical biotinylation: over-biotinylation is excluded, ideally as assessed by mass spectrometry.
-
14.
If possible, the biotinylated target protein is supplemented with 10% glycerol, aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C. To assess if freezing is tolerated by the target protein, compare a frozen/thawed and untreated sample by SEC. If no discernible aggregation or protein loss is observed, freezing can be considered as tolerated.
-
15.
We routinely freeze biotinylated membrane proteins for storage purposes. Having thus far analyzed more than a dozen membrane proteins in this manner, we never experienced aggregation problems due to freezing. Frozen biotinylated proteins remain stable at −80 °C for several years.
-
16.
The preferred location of the Avi tag on the target protein depends on the quality and quantity of the fusion protein that can be obtained. Both parameters are most easily assessed by using fluorescence-detection size-exclusion chromatography (FSEC) analysis [25].
-
17.
Biotinylation of Avi-tags during cultivation can be achieved in several additional expression systems [14, 26, 27]. These procedures also require the co-expression of BirA and growth medium supplemented with biotin.
-
18.
Numerous expression screenings provided the tendency that Expi293 is more successful for expressing membrane proteins. As an alternative we recommend Freestyle 293-F cells. Implementation of the latter will require small adaptations of the described workflow for which we refer to the instructions from the supplier.
-
19.
Supplementing the medium with biotin is optional. Over the course of many years and targets we observed virtually complete biotinylation even in the absence of supplemented biotin.
-
20.
It is very important that compounds containing primary amines are absent from the purified protein sample for chemical biotinylation. A frequent source of primary amines stems from Tris-buffers. IMAC-purified protein is not pure enough regarding biogenic amines to be used for NHS coupling.
-
21.
The NHS moiety of EZ-Link Sulfo-NHS-LC-Biotin reacts with water and is thereby inactivated. We therefore highly recommend preparing the Sulfo-NHS-LC-Biotin solution freshly. Keep solid EZ-Link Sulfo-NHS-LC-Biotin under argon at −80 °C for prolonged storage.
-
22.
In case the target protein is unstable at 25 °C, the biotinylation reaction can be carried out at 4 °C. In this case, increase the biotin-target protein ratio to 10:1 and incubate for 60 min instead of 30 min.
-
23.
A typical pattern contains different species containing either none, one, or several biotin moieties per target protein (Fig. 1). Ideally, the non-biotinylated species should not exceed 10% of the total species (in the example of GFP labeling shown in Fig. 1, non-biotinylated target accounts for around 5%). In case of over- or under-biotinylation, the biotin-target protein ratio needs to be adjusted accordingly, while keeping the target protein concentration and incubation time constant.
-
24.
The biotinylation of target protein can be quantified by mobility shift in SDS-PAGE upon addition of streptavidin to the sample. Streptavidin remains folded and bound to the biotinylated target protein under conventional SDS-PAGE conditions [28]. Membrane protein samples are usually not boiled before SDS-PAGE. However, when boiling the sample is required add streptavidin afterward.
-
25.
Due to the tetrameric architecture of streptavidin with four biotin-binding sites, multiple protein bands may be observed, e.g., (1) free streptavidin (53 kDa), (2) streptavidin associated with a single target protein, and (3) streptavidin associated with multiple (up to four) target proteins. In our hands, it is more straightforward to use the intensity loss of the target protein band upon streptavidin addition relative to the control sample for quantification. For more precise quantification we recommend mass spectrometry to analyze the degree of biotinylation. For qualitative analysis of target protein biotinylation, western blotting using a streptavidin-HRP conjugate can be employed.
-
26.
Incomplete biotinylation might be advantageous regarding oligomeric proteins. Similar to the presence of multiple biotin labels on monomeric proteins, the occurrence of multiple biotin groups per oligomeric protein complex may restrict its flexibility upon immobilization and thereby decrease the variation and amount of protein surface accessible to the binders. We recommend a pull-down of biotinylated target protein with immobilized streptavidin and compare the pull-down efficiency with the mobility shift in SDS-PAGE to quantify the biotinylation per oligomeric unit.
References
Steyaert J, Kobilka BK (2011) Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol 21:567–572
Zimmermann I, Egloff P, Hutter CA et al (2018) Synthetic single domain antibodies for the conformational trapping of membrane proteins. elife 7
Rusmini F, Zhong Z, Feijen J (2007) Protein immobilization strategies for protein biochips. Biomacromolecules 8:1775–1789
Fairhead M, Howarth M (2015) Site-specific biotinylation of purified proteins using BirA. Methods Mol Biol 1266:171–184
Green NM (1990) Avidin and streptavidin. Methods Enzymol 184:51–67
Piran U, Riordan WJ (1990) Dissociation rate constant of the biotin-streptavidin complex. J Immunol Methods 133:141–143
Marttila AT, Laitinen OH, Airenne KJ et al (2000) Recombinant NeutraLite avidin: a non-glycosylated, acidic mutant of chicken avidin that exhibits high affinity for biotin and low non-specific binding properties. FEBS Lett 467:31–36
Green NM (1975) Avidin. Adv Protein Chem 29:85–133
Sano T, Cantor CR (1990) Cooperative biotin binding by streptavidin. Electrophoretic behavior and subunit association of streptavidin in the presence of 6 M urea. J Biol Chem 265:3369–3373
Fall RR (1979) Analysis of microbial biotin proteins. Methods Enzymol 62:390–398
Robinson BH, Oei J, Saunders M et al (1983) [3H]biotin-labeled proteins in cultured human skin fibroblasts from patients with pyruvate carboxylase deficiency. J Biol Chem 258:6660–6664
Beckett D, Kovaleva E, Schatz PJ (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci 8:921–929
Schatz PJ (1993) Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 11:1138–1143
Cull MG, Schatz PJ (2000) Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol 326:430–440
Brown MT, Steel BC, Silvestrin C et al (2012) Flagellar hook flexibility is essential for bundle formation in swimming Escherichia coli cells. J Bacteriol 194:3495–3501
Guzman LM, Belin D, Carson MJ et al (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130
Hartley JL, Temple GF, Brasch MA (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10:1788–1795
Casadaban MJ, Cohen SN (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol 138:179–207
Howarth M, Takao K, Hayashi Y et al (2005) Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci U S A 102:7583–7588
Ullah R, Shah MA, Tufail S et al (2016) Activity of the Human Rhinovirus 3C Protease Studied in Various Buffers, Additives and Detergents Solutions for Recombinant Protein Production. PLoS One 11:e0153436
Don RH, Cox PT, Wainwright BJ et al (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Marino J, Holzhuter K, Kuhn B et al (2017) Efficient Screening and Optimization of Membrane Protein Production in Escherichia coli. Methods Enzymol 594:139–164
Schmidt TG, Skerra A (1994) One-step affinity purification of bacterially produced proteins by means of the “Strep tag” and immobilized recombinant core streptavidin. J Chromatogr A 676:337–345
Keefe AD, Wilson DS, Seelig B et al (2001) One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr Purif 23:440–446
Kawate T, Gouaux E (2006) Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14:673–681
Erdmann D, Zimmermann C, Fontana P et al (2010) Simultaneous protein expression and modification: an efficient approach for production of unphosphorylated and biotinylated receptor tyrosine kinases by triple infection in the baculovirus expression system. J Biomol Tech 21:9–17
Thie H, Binius S, Schirrmann T et al (2009) Multimerization domains for antibody phage display and antibody production. New Biotechnol 26:314–321
Viens A, Mechold U, Lehrmann H et al (2004) Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal Biochem 325:68–76
Acknowledgments
We thank Martin Siegrist, Jean-Marie Vonach, and Marcello Foggetta for technical help during expression and fermentation as well as Roche Diagnostics in Munich, Penzberg for reagent generation. We acknowledge financial support from the German Research Foundation via the Cluster of Excellence Frankfurt (Macromolecular Complexes, to E.R.G.), and the SFB807 (Transport and Communication across Biological Membranes, to E.R.G.). Work in the Seeger group was supported by a SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823, to M.A.S.), a SNSF NRP 72 grant (407240_177368, to M.A.S.), a SNSF BRIDGE proof-of-concept grant (20B1-1_175192, to P.E.), and a BioEntrepreneur-Fellowship of the University of Zurich (BIOEF-17-002, to I.Z.). R.J.P.D, E.R.G., and M.A.S. acknowledge a grant of the Commission for Technology and Innovation CTI (16003.1 PFLS-LS).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this protocol
Cite this protocol
Kuhn, B.T. et al. (2020). Biotinylation of Membrane Proteins for Binder Selections. In: Perez, C., Maier, T. (eds) Expression, Purification, and Structural Biology of Membrane Proteins. Methods in Molecular Biology, vol 2127. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0373-4_11
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0373-4_11
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0372-7
Online ISBN: 978-1-0716-0373-4
eBook Packages: Springer Protocols