Abstract
The ascomycete filamentous fungus Neurospora crassa played a historic role in experimental biology and became a model system for genetic research. Stimulated by a systematic effort to collect wild strains initiated by Stanford geneticist David Perkins, the genus Neurospora has also become a basic model for the study of evolutionary processes, speciation, and population biology. In this chapter, we will first trace the history that brought Neurospora into the era of population genomics. We will then cover the major contributions of population genomic investigations using Neurospora to our understanding of microbial biogeography and speciation, and review recent work using population genomics and genome-wide association mapping that illustrates the unique potential of Neurospora as a model for identifying the genetic basis of (potentially adaptive) phenotypes in filamentous fungi. The advent of population genomics has contributed to firmly establish Neurospora as a complete model system and we hope our review will entice biologists to include Neurospora in their research.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Ascomycete
- Filamentous fungi
- Population genomics
- Biogeography
- Speciation
- Reverse ecology
- Introgression
- Self–nonself recognition
- Selective sweep
1 Introduction: Fungi and Population Genomics
Among complex eukaryotes, fungi have excellent potential as models for population studies at diverse levels, and in particular at the genomic level [1,2,3]. Population genetics as a discipline has long been largely concerned with plants and animals [4], but this trend is currently being tempered by the massive production of fungal genomic data. The first eukaryote to have its genome sequenced was fungal (the baker’s yeast Saccharomyces cerevisiae) and the rate of genomic sequences production is higher in the fungal kingdom than in any other eukaryotic kingdom. For instance, as of mid-2017, an estimated 2000 fungal genomes have been sequenced and assembled, and several thousand resequenced genomes are available for population genomic investigations [5]. Fungi have relatively small and low-complexity genomes by eukaryotic standards (typically 30–40 Mb, ~10,000 genes), many fungi have haploid genetics, and these genomic advantages have contributed to making fungi the leading kingdom for eukaryotic genome sequencing [6]. It also follows that fungal population genomics is the only variety of eukaryotic population genomics that is truly “genomic,” given that, unlike fungi, most plant and animals genomes cannot be sequenced telomere-to-telomere in relatively large numbers within a reasonable time. However, in the context of evolutionary and ecological genetics, what has long been lacking is access to—and essential information on—fungal natural populations [4]. The genus Neurospora stood out early as an outstanding model for fungal population studies, with large numbers of isolates that could be sampled in a predictable manner in various ecosystems [2]. In this chapter we will begin by briefly summarizing the biological features, human and historical factors that have contributed to bring Neurospora into the realm of evolutionary biology and ecology. We will then cover the major contributions of Neurospora to our understanding of fungal biogeography, fungal speciation and the permeability of barriers to gene flow. Finally, we will review the early contributions of Neurospora to our current knowledge of the genetic basis of (potentially adaptive) phenotypes in filamentous fungi.
2 The Rise of Neurospora as a Model for Evolutionary and Ecological Genetics
Neurospora is one of the most easily recognized of filamentous ascomycetes (Fig. 1). Originally described as a contaminant in French army bakeries [7], Neurospora is most often encountered as powdery masses of bright, carotenoid-colored mycelium and mitospores (=conidia, see Note 1) on the surface of burned or heated substrates. Visible, pink to orange colonies on scorched vegetation or cooked foodstuffs form the primary source of Neurospora collections, but aconidial noncolored species can also be isolated from heat-treated soil. The ecological components of the life cycle of Neurospora are not fully understood and might involve close association with plants, such as endophytism [8]. Sex, however, is well understood, beginning with the discovery of sexual fruiting bodies (perithecia, see Note 1) with meiotic products aligned in linear tetrads by mycologists Cornelius Shear and Bernard Dodge [9]. These ordered tetrads stimulated the use of Neurospora as the fungal rival of Drosophila and maize as a model for genetic research [10,11,12]. The fact that Neurospora is an haplont (which facilitates recognition of recessive loss-of-function mutations) and that it can be grown on simple minimal media (making it possible to impose further nutritional requirements by mutation) were other salient biological features that popularized Neurospora for genetic investigations [12, 13]. In 1941, George Beadle and Edward Tatum used Neurospora to obtain the first biochemical mutants and to show that genes control metabolic reactions, which was referred to as the “one gene–one enzyme” hypothesis [14, 15]. Beadle and Tatum’s experiments helped “convince many skeptical biologists that genes control the fundamental processes of life, and not just the final touches of development, such as wing shape or eye pigment” and “started a new era by bringing genetics and biochemistry together” [13].
In parallel with the adoption of Neurospora as a model for molecular and cell biology, David Perkins introduced Neurospora into the realm of evolutionary biology and ecology in 1968 by putting in place the long-term study of wild populations [11]. The initial objective of the systematic sampling initiative set up by Perkins was to provide genetic variants for laboratory investigations. Although the mode of primary colonization of Neurospora and other fundamental aspects of the ecology of these organisms remain a particular mystery, a distinctive advantage of Neurospora relative to many microbes was the relative ease of sampling in diverse ecosystems [2, 16]. Perkins, Dave Jacobson and other scholars eventually gathered a collection of >5000 isolates, access to which is still provided by the Fungal Genetics Stock Center (University of Kansas). Surveys of the wild strains have continued to provide genetic variants for a variety of laboratory investigations on mitochondria and senescence plasmids, genes governing vegetative incompatibility or mating types, and meiotic drive or transposable elements [4, 17, 18]. Neurospora entered the genomic era in 2003 with the release of its genomic sequence [19], and the Perkins collection was quickly perceived as a boon by functional, ecological, and evolutionary genomicists. Recently, wild isolates proved valuable for variation-guided functional analyses of cell–cell recognition, via either genome-wide association studies or QTL mapping [20,21,22]. Resequencing data for wild isolates has also been used to identify genes that underlie variation in phenotypes related to self-recognition and cold tolerance [22, 23]; reviewed below in the final section 5). More generally, the collection also advanced knowledge of the systematics, biogeography, population biology, and evolutionary history of Neurospora and ascomycetes in general [2, 17, 24]; reviewed in next two sections 3 and 4).
3 Neurospora Population Genomics Has Revealed Cryptic Species with Large Variation in the Extent of Their Geographical Distribution
3.1 Nothing Is Generally Everywhere
Microbes have long been thought to have large geographic distributions, in contrast to the highly restricted ranges of larger organisms. Fungi were not immune to this misconception, and the idea that dispersal ability per se does not limit the geographic distribution of these organisms remains quite widespread today, even among biologists. The misconception that many fungi had global distributions is largely based on two factors. First, the observation that almost all fungi produce tiny, powder-like propagules on structures promoting their dissemination by wind [32] and second, reliance on morphological species recognition criteria that have proved to be too broad for fungi, and have given an inaccurate picture of fungal diversity, distributions and ecologies [33, 34].
3.2 Geographic Endemicity Within Globally Distributed Neurospora Morphospecies
Studies on wild Neurospora isolates have altered our understanding of fungal biogeography, providing a perfect illustration that the inferred geographic range of a fungal species depends upon the method of species recognition. More generally, studies on wild Neurospora isolates have shown that fungal species are highly structured and that fungal distribution have been shaped by geological and climatic events the same way as macrobes have [34, 35]. Conventional criteria based on morphology are of little use to ascertain taxonomic status in Neurospora as most conidiating species cannot be distinguished from one another by the size, color and shape of their vegetative and reproductive organs [9, 16, 17]. Hence, by morphological species recognition, only two species of conidiating Neurospora are found: one with eight ascospores per ascus, and one with four ascospores per ascus (see Note 1). The two morphospecies are both cosmopolitan in temperate and tropical latitudes (Table 1). In vitro mating compatibility tests and phylogenetic analyses, however, reveal that the two morphological species have their own biogeography and encompass multiple endemic species: under biological species recognition seven species are found, while under phylogenetic species recognition at least twenty-six species are identified (Table 1). Similar to many plant and animal genera, the distribution of some of the phylogenetic species appears to be quite limited (e.g., N. hispaniola was reported in the Hispaniola island only) while others have very broad distributions (e.g., globally distributed N. discreta PS4). Genealogies of multiple genes have been widely used in fungi, including Neurospora [24, 25, 29,30,31], improving our understanding of the structure of fungal biodiversity. The observation that fungal species defined by morphology typically harbor several to many endemic species can be explained by the relative paucity of morphological characters and the slower rate of morphological change for organisms with less elaborate development and fewer cells, allowing genetic isolation to precede recognizable morphological changes [34]. Just as genetic isolation can precede morphological change, phylogenetic divergence can precede reproductive isolation, such that one biological species can embrace several phylogenetic species, as was shown when biological and phylogenetic species recognition were compared in Neurospora [36]. Species recognition by genealogical concordance, popular as it has been, has limitations, related to heterogeneity in the congruence of sequenced loci with the species tree, and inadequate sampling of substrates throughout the geographic range of species; Neurospora is not an exception.
3.3 On the Difficulty of Species Diagnosis in Neurospora and Fungi
Morphology is of little use to identify species in Neurospora, like in many microscopic filamentous fungi. Perkins and collaborators have published a bountiful collection of protocols to induce and assess mating in vitro in Neurospora, but systematic analyses of pre- and postmating barriers in large collection of isolates is challenging to implement in modern labs. Species recognition based on genealogical concordance among gene trees became the gold-standard in Neurospora, revealing cryptic species diversity [34]. Species recognition by genealogical concordance, however, suffers from two limitations. The first limitation is the requirement for the use of the same sets of sequenced loci across studies. The second limitation is that the resolving power of sequenced loci is most often not known a priori, and markers not chosen based prior knowledge of the species tree.
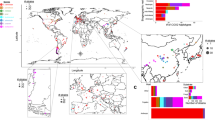
Figure 2 provides an illustration of the phylogenetic diversity encompassed by the genus Neurospora, but also of the limitations of species recognition by genealogical concordance. Sequences at six loci previously identified by Nygren et al. [38] were retrieved from GenBank, extracted from publicly available genomes [22, 28, 39] and extracted from the genomes of isolates originating from multiple sites in North America (De Bellis et al., unpublished) and isolated from the soil spore bank treated with heat (Hann-Soden et al., unpublished). Sequences at the six loci were concatenated and the resulting total evidence tree recapitulates the phylogenetic species identified by applying species recognition by genealogical concordance sensu Dettman et al. [29]. Two important insights emerge from this analysis. The first striking feature is that the species identified are different from the species previously identified by Dettman et al. [29] using three loci not included in the Nygren et al. [38] loci. Many of the phylogenetic and biological species previously identified do not stand as distinct species in this analysis. Another important result is the relatively large number of species that can coexist within the same spore bank (not shown here). In summary, using half a dozen loci for phylogenetic species delineation makes it more operational but also limits its ability to resolve population subdivision. Genomic data should have increased power to resolve species limits and evolutionary relationships, but standard species recognition by genealogical concordance remains useful until the cost of sequencing a set of markers is higher than sequencing a full genome.
Total evidence genealogy inferred using RAxML v8 [37] based on the concatenation of sequences at six loci published by [38], including Bml, mak-2, nik-1, ORF1, pkc, and tef-1. Phylogenetic species identified by genealogical concordance across multiple gene genealogies are indicated by shaded areas and bootstrap supports in the total evidence phylogeny. Taxon names are reported only for best-sampled conidiating species. Nhis, N. hispaniola; Nsit, N. sitophila; Nper, N. perkinsii; Nint, N. intermedia; Nmet, N. metzenbergii. Marker sequences were retrieved from genomic sequences using blastn for all datasets but the ref. [38] dataset which was downloaded from NCBI. Samples from Hann-Soden et al. (Unpublished) and some of the samples from Nygren et al. [38] were isolated from heat-treated soil, while the remaining samples were isolated from burned vegetation
3.4 Population Structure Within Neurospora Phylogenetic Species
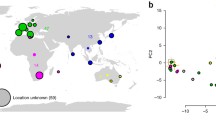
Phylogenetic analyses and mating compatibility tests have great potential to augment knowledge of the taxonomy, ecology and biogeography of fungal genera and species, but these approaches are not operational when the goal is to infer fine-scale population genetic structure [34, 40]. High-throughput sequencing technologies have made it possible for individual laboratories to acquire whole-genome sequences across populations and test hypotheses of geographic endemicity or genome evolution previously formulated based on sequence diversity and reproductive biology. Population genomic studies of eight-spore biological species of Neurospora are illustrative of the finer resolution afforded by genomic information to characterize population structure. For instance, in selecting populations of Neurospora for genome wide association studies, researchers relied on populations previously identified by concordance of gene or microsatellite genealogies that included a strain with a reference genome and that spanned significant environmental variation. In the case of N. crassa, the population boarded the Gulf of Mexico and for N. discreta PS4, the population displayed a remarkably large latitudinal distribution along western North America. However, in N. crassa phylogenomic analyses and model-based Bayesian clustering of transcriptomic data for 50 isolates revealed not 1 population, but multiple divergent lineages, with the two best sampled lineages found in Louisiana and the Caribbean [23] (Fig. 3). Subsequent phylogenomic analyses of SNPs from the resequenced transcriptomes of 112 N. crassa individuals from the same geographic area as the Louisiana population showed no population subdivision, providing an ideal setting for a genome-wide association study (see last section). In the same way, phylogenomic analyses and model-based Bayesian clustering of whole genome information for 128 N. discreta PS4 isolates revealed not one population, but six divergent lineages [39] (De Bellis et al. unpublished) (Fig. 3). Demographic inference based on a diffusion approximation to the site frequency spectrum [46] estimated relatively recent divergence times (≈0.4 M generations) between the lineages discovered within N. crassa and N. discreta PS4 [23, 39]. Relating these divergence time estimates with historical events requires a generation time, which is a perennial question in Neurospora, and in fungi in general. Extensive observations of Neurospora colonies at burn sites led to only a few observations of fruiting bodies in nature, possibly owing to the difficulty in the recognition of black perithecia on/within burned substrate, or the delay of sexual reproduction after conidial blooms [18]. Perithecia were observed on maize cobs [47], under the bark of fire-injured trees [2, 48], or protruding through cracked tissues of scorched sugar cane [49]. However, it remains unknown whether sexual cycles in Neurospora are synchronized with wildfires. As proposed by Turner et al. [18], “Perhaps the dramatic conidiating blooms seen on burned or scorched vegetation are exceptional sporadic events punctuating a mode of growth that is otherwise inconspicuous or invisible.” Given these uncertainties, a plausible scenario is that divergence within North American N. crassa and N. discreta PS4 (≈0.4 M generations) has been driven by climate oscillations of the Pleistocene (2.6 Mya–11 kya), but alternatives cannot be ruled out without further information on the generation time and other aspects of the population biology of Neurospora.
(a) Population genomic structure of Neurospora discreta PS4 as inferred from whole genome resequencing of 128 isolates (ref. [39] and De Bellis et al., Unpublished). Left: RAxML8 whole genome genealogy; center: ancestry proportions in K = 7 clusters as inferred using sparse Nonnegative Matrix Factorization [41]; right: sampling sites in North America and proportions of isolates assigned to four European/North American clusters as pie charts. (b) Population genomic structure of American Neurospora crassa as inferred from reanalysis of previously published transcriptome resequencing data [23]. Left: Neighbor-Net inferred from biallelic SNPs without missing data using Splitstree [42]; center: ancestry proportions in K = 4 cluster as inferred using Structure 2.3.4 [43,44,45] based on a random 10% of the 134 k SNPs without missing data; right: sampling sites in America and sum of membership proportions in four Structure clusters as pie charts. US/Mexico State Abbreviations: AK Alaska, CA California, FL Florida, ID Idaho, LA Louisiana, MT Montana, NV Nevada, WA Washington, YU Yucatan. ISO country codes: CH Switzerland, CI Côte d’Ivoire, CR Costa Rica, ES Spain, GY Guyana, HA Haiti, PA Panama, PG Papua New Guinea, PT Portugal, TH Thailand, VE Venezuela
3.5 Comparative Population Genomics of Selfing and Outcrossing Neurospora Species
Like plant evolutionary biologists, the evolutionary causes and consequences of self-fertilization is a question of long-standing interest to research scholars working with Neurospora [50, 51]. It was recognized at an early stage that the variety of lifestyles in Neurospora shows promise for comparative studies [12]. In contrast to the eight-spored Neurospora species, meiotic products in the morphospecies N. tetrasperma are packaged into four relatively large ascospores [52]. In nature and in the laboratory, the mycelium that emerges from germinating N. tetrasperma ascospores is normally heterokaryotic with component nuclei of opposite mating type (A and a; see Note 2). Neurospora tetrasperma therefore superficially resembles true homothallic (see Note 3) species [53] in that each ascospore can usually produce a self-fertile mycelium and complete the sexual cycle without needing to find a compatible mate [52]. As a consequence, the breeding system (see Note 4) of N. tetrasperma is referred to as “pseudohomothallic” (see Note 3) [54]. Such reproductive systems (see Note 4) based on heterokaryosis (see Note 5) are unique to fungi, and they can be described as “a form of heterothallism with provisions to allow prolonged inbreeding” (see Note 3) [55]. In the laboratory, heterokaryotic (see Note 5) wild-type strains self, but self-sterile homokaryotic (A or a) conidia (see Note 5) derived from wild-type strains are functionally heterothallic (see Note 3), and can be outcrossed [4, 17, 52, 56]. Recently the frequency of homokaryotic conidium production by pseudohomothallic N. tetrasperma strains collected from New Zealand and the UK was systematically studied [27]. Large differences in the number of homokaryotic conidia produced by different populations were observed, with the rate of homokaryotic conidia being twice as high as previously reported [56], suggesting ample opportunities for outcrossing sex. Laboratory crosses employing homokaryotic isolates of opposite mating types obtained from different strains, however, revealed a high frequency of sexual dysfunction caused by vegetative incompatibility between interacting mycelia [55, 57]. These mating compatibility studies suggest that outcrossing may be limited in nature and that large difference in self-sterile spore production across N. tetrasperma populations is not necessarily associated with large difference in outcrossing rates.
More recently, population genomics was used to test if vegetative incompatibility is effectively blocking outcrossing in N. tetrasperma populations, by quantifying the level of outcrossing in situ and correlating this factor to population structure and genome evolution [28]. Phylogenomic analysis and model-based clustering of whole genome information for large set of strains confirmed nine of the ten cryptic phylogenetic species previously identified [25,26,27] and revealed an additional lineage in Europe, but no subdivision within species was detected [28, 58]. Nucleotide diversity was of the same order of magnitude in populations of N. tetrasperma as seen in populations of outcrossers N. crassa and N. discreta, suggesting no strong reduction of within-population diversity in N. tetrasperma as would be expected under inbreeding (Fig. 4). Analyses of linkage disequilibrium (Chapter 1) were consistent also consistent with selfing [28] (Fig. 4). The ratio of nonsynonymous to synonymous nucleotide diversity (πN/πS) was relatively high in N. tetrasperma (>0.7), suggesting a relatively high proportion of slightly deleterious mutation consistent with selfing, although πN/πS ratios were comparable to those observed in the outcrosser N. crassa (Fig. 4). There is a difference between the 8-spored, heterothallic and 4-spored pseudohomothallic species, however, when it comes to species recognition by concordance of gene genealogies. With N. discreta and N. crassa, 400 k year old lineages were not diagnosed as species, but the 700 k year old lineages (not appreciably from 400 k in this instance) within N. tetrasperma were diagnosed as distinct phylogenetic species. This difference is consistent with theoretical predictions, because population differentiation should increase as consequence of lower within-deme diversity and the combined action of other reproductive and life-history traits which tend to increase isolation [61].
Summary statistics of population genomic variation in best-sampled Neurospora lineages. For N. discreta unpublished whole-genome resequencing data (De Bellis et al., Unpublished) were aligned against the reference genome FGSC8579. For N. crassa, transcriptome resequencing were downloaded from Short Read Archive (accession SRA026962; [23]) and aligned against reference OR74A v2.0 (hosted at Ensembl Fungi). For N. tetrasperma, VCF files were downloaded from the Dryad Digital Repository (https://datadryad.org/resource/doi: https://doi.org/10.5061/dryad.162mh; [28]). LD decay values for N. tetrasperma and N. crassa were previously published by [23, 28]. Computations were carried out in Egglib v3 [59], excluding codon-coding nucleotide triplets with missing data. πS is the nucleotide diversity at synonymous sites. LD-decay 50% is the distance over which linkage disequilibrium (LD) decays to half its maximum. For N. tetrasperma L8 and L10, actual values of LD-decay 50% are 31 kb and >500 kb and bars were truncated for clarity for these two lineages. πN/πS is the ratio of nonsynonymous to synonymous nucleotide diversity, which gives, under near neutrality, an estimate of the proportion of effectively neutral mutations that are strongly dependent on the effective population size Ne [60]. The mating-type chromosome of N. tetrasperma was excluded from calculations. Ndis NW Northwestern Neurospora discreta lineage (purple in Fig. 2), Ndis SW Southwestern Neurospora discreta lineage (orange in Fig. 2), Ndis SE Southeastern Neurospora discreta lineage (dark green in Fig. 2), Ndis PG Neurospora discreta lineage from Papua New Guinea (light blue in Fig. 2), Ntet L5 Neurospora tetrasperma lineage 5, Ntet L8 Neurospora tetrasperma lineage 8, Ntet L10 Neurospora tetrasperma lineage 10, Ncra LA Neurospora crassa Louisiana lineage, Ncra CAR Neurospora crassa Caribbean lineage
4 Neurospora Population Genomics Has Refined Our Views on the Permeability of Barriers to Gene Flow
Population genomics offers great potential for enhanced characterization of cryptic population subdivision in Neurospora, revealing that the genus, which is ubiquitous in temperate, subtropical and tropical regions, is structured into a variety of species and divergent lineages within species. The observed richness in Neurospora species and lineages does not result in complete geographic separation (i.e., allopatry) and many lineages or species have overlapping ranges (i.e., sympatry or parapatry) (Table 1). Not only multiple species [4, 62], but also multiple lineages within species [39] (Fig. 3), may be collected from the same sites, just centimeters from each other, raising the question of hybridization and admixture [63, 64]. Although true hybrids have not been found in nature [29], the production in the lab of small but significant numbers of viable hybrid progeny has been known since the description of the genus ([65] cited by [58]) and repeatedly confirmed since then (e.g., [18, 36, 66]). For instance, crosses between allopatric N. intermedia and N. crassa in the lab can typically yield 1–15% black (i.e., potentially viable) ascospores (see Note 1) [36]. The problem with studying hybridization in Neurospora and in fungi in general has been that neither a discerning eye nor simple mating tests can reliably identify hybrids among strains in situ; this is where genomic approaches come to the rescue of the mycologist.
Genomic studies of Neurospora populations provided indirect evidence for hybridization and introgression. The first element supporting the existence of hybridization came from the outcrossing heterothallic N. crassa (see Note 3). A genomic island of elevated relative and absolute divergence identified between the Louisiana and Caribbean populations of N. crassa [23] showed an unusually large number of fixed differences, suggesting that divergence between haplotypes was older that the splitting of populations. Haplotypic structure at the genomic island was also different between the two populations, with less haplotype diversity and nonuniform haplotype boundaries in the Louisiana population. Together these observations point to the introgression and selective sweep of a single migrant tract in Louisiana from a more genetically diverged population or species that remains to be identified. Further indirect evidence for hybridization came from comparative and population genomic investigations of the pseudohomothallic N. tetrasperma (see Note 3), providing a good illustration of the impact of mating system on population structure in fungi. As described in the previous section, the meiotic pathway of N. tetrasperma was reprogrammed so that each ascospore receives mat A and mat a haploid nuclei produced from a single diploid nucleus, which favors selfing. This particular meiotic process is dependent on the segregation of mating-type alleles at the first division of meiosis, which is assured by the suppression of recombination between the centromere and the mating-type locus [67]. Because recombination is suppressed, a large part of the mating-type chromosomes of N. tetrasperma degenerates, accumulating nonsynonymous polymorphisms/substitutions and nonoptimal codons [28]. However, genome-wide analyses of N. tetrasperma revealed patterns of divergence consistent with a history of introgressive hybridization with several heterothallic relatives, and in particular large tracts of DNA of allospecific origin restricted to the mating-type chromosome. It is also worth noting that these hybridization events occurred despite the morphologically enforced, preferentially selfing, mating system of N. tetrasperma. The introgressed tracts have been fixed within N. tetrasperma lineages and some of them carry signatures of selective sweeps, suggesting that they confer an adaptive advantage in natural populations. An hypothesis is that the introgression of nondegenerated mating type chromosomes from related species may contribute to maintain integrity of mating type chromosome and constitute a process of genomic reinvigoration acting to reduce the mutational load. These findings corroborate a prediction made by Metzenberg and Randall, and reported by ref. [18] as a personal communication: “periodically during evolution, a deteriorated mating type chromosome is replaced following a cross between N. tetrasperma and one of the heterothallic species.”
The population genomic studies presented so far show evidence of introgressive hybridization, but none has caught populations in the act of mixing. The most recent evidence for population admixture comes from the study of the population genomic structure of a single species, the outcrossing N. discreta PS4. Phylogenomic analyses and model-based clustering of whole-genome data showed four, well-diverged lineages: Papua New-Guinea (PNG), Alaska and Europe (AK-EU), California and Washington state (CA-WA), and New Mexico and Washington state (NM-WA). Admixture analyses using Frappe and a genome scan for lineage-diagnostic SNPs revealed an Alaskan strain that possesses 12% of the genome of the apparently allopatric, NM-WA lineage [39]. Yet, at the same time, there was no evidence of admixture at one collecting site in Washington where the CA-WA and NM-WA lineages are clearly sympatric. The finding of one admixed individual suggested that reproductive isolation is not complete between all pairs of lineages within N. discreta and that there might have been opportunities for gene flow between them. Analyses of postdivergence gene flow using the dadi package [46], which infers demographic parameters based on a diffusion approximation to the site frequency spectrum, supported models of gene flow following secondary contact, both between North American and PNG lineages, and among lineages in North America/Europe. The finding of nonzero migration rates between all lineages suggests that their geographic distributions have been overlapping to some extent. In North America/Europe, lineages may have diverged following repeated periods of isolation in separate glacial refugia [68], interspersed with periods of secondary contact potentially permitting gene flow that only began relatively recently, as suggested by the finding of a late generation hybrid in Alaska indicative of ongoing admixture.
The best supported models of secondary contact between North American/European lineages assumed heterogeneous gene flow across the genome, and parameter estimates indicated that only a small fraction of the genome experienced relatively higher migration rates between lineages. This heterogeneity in introgression rates may reflect the indirect effects of selection, with limited gene flow in the neighborhood of barrier loci (genetic incompatibilities or genomic features under divergent ecological selection), higher gene flow at adaptively introgressed regions, and basal gene flow in regions not affected by selection [69]. The widespread compatibility of crosses attempted in vitro between North American/European indicated a lack of intrinsic premating barriers (i.e., assortative mating by mate choice), intrinsic postmating prezygotic barriers (i.e., gametic incompatibility) or a form of intrinsic early postzygotic isolation (i.e., zygotic mortality), and the absence of these barriers may have contributed to facilitate gene flow following secondary contact. The lack of intrinsic premating barriers appears to be general among Neurospora lineages and species, and may result from constraints on the evolution of pheromone-receptor systems involved in mating [70]. The peptide pheromones that mediate premating attraction between mating-type compatible isolates are identical between Neurospora and outgroups [71], consistent with a lack of pheromone-based mate choice. Together these findings and observations suggest that the barriers that limit gene flow following secondary contact and account for the observed heterogeneity in migration rates are mostly extrinsic barriers (e.g., immigrant inviability) or late intrinsic barriers (e.g., hybrid inviability or sterility). This work also illustrates the great potential of speciation genomics for increasing our understanding of fungal biogeography, revealing features such as sympatry in the recent past or admixture between apparently allopatric species, that would not be accessible without genomic data, especially given the scarcity of exploitable fossil records in fungi. Further work on the fine-scale population genetic structure of Neurospora is required to quantify lineage diversity at the local scale and the extent of interlineage admixture, and more detailed investigations of barriers to gene flow (e.g., by measuring hybrid inviability and sterility) should provide more insights on the factors contributing to the maintenance or mixing of lineages in sympatry.
5 Studies Neurospora Provide Insights into the Genetic Basis of (Potentially Adaptive) Phenotypes in Wild Microbial Eukaryotes
The difficulty of defining the boundaries of populations has been a major impediment for studying the genetic basis of differences between individuals within populations and of adaptive differences between populations. However, by revealing geographic endemism, genomic approaches to characterizing fungal populations have offered new opportunities to identify the molecular underpinnings of key biological features in Neurospora. In cases where segregating phenotypic traits could be scored in a Neurospora population, quantitative trait locus (QTL) mapping or genome-wide association studies (GWAS) could be used successfully to identify the underlying genomic features. The rates of LD decay observed in N. crassa or N. discreta for instance (Table 1) are suitable for linkage mapping. QTL mapping was used to identify loci in N. crassa associated with a reinforced, post-mating, female mate choice barrier between N. crassa and N. intermedia [72]. However, due to the presence of large regions of linkage inherent to the QTL approach, the authors could not identify the genes responsible for the quantitative trait. When combined with dense, genome-wide marker coverage and lack of population structure, GWAS can overcome the limitations of QTL studies and offer higher resolution by increasing the range of genetic and phenotypic variation surveyed, by avoiding the generation of time-consuming crosses and taking advantage of many more generations of recombination. GWAS has been used successfully in N. crassa to identify the genetic basis of the complex trait of germling communication, the process by which conidia germinating near each other can sense each other, reorient their growth toward one another, and fuse (see Note 6). By quantifying the proportion of communicating germlings in germinating populations of conidia representing 24 Louisiana N. crassa isolates, and using RNAseq to genotype isolates, Palma-Guerrero et al. [21] successfully associated a calcium sensor with the cell-to-cell communication trait.
Another strategy used for rapid trait mapping in segregating populations or crosses is bulked segregant analysis, that is, the genotyping or sequencing of bulked pools of segregating individuals with the most extreme phenotypes. The advantage of bulked segregant analysis over standard QTL analysis or GWAS is that there is no necessity for genotyping all individuals in the segregating population or progeny, which can increase the number of progeny surveyed and hence decrease the size of linked regions surrounding quantitative trait loci compared to classical approaches. As a proof of principle for mutation mapping by bulked segregant analysis, Pomraning et al. [73] used bulked segregant analysis to map the mutation(s) underlying temperature-responsive cell cycle regulation in the classic N. crassa ndc-1 (nuclear division cycle-1) mutant. Two hundred progeny from a wild x mutant cross were tested for the arrest of the nuclear division cycle just prior to DNA synthesis when grown at 37 °C, and a subset of 63 progeny with extreme phenotypes was sequenced en masse in two pools, allowing identification of a single mutation in a single gene.
In cases where bulked segregant analysis does not pinpoint a single gene, population genomic approaches can be used to identify the genes associated with traits. This approach has been demonstrated by studies of the genetic basis of cell communication and fusion in Neurospora (see Note 6). Heller et al. [20, 74] used bulked segregant analysis to map the genes responsible for kind discrimination among germlings and rapid cell death following germling fusion (i.e., “germling regulated death”), identifying 100 kb and 180 kb regions of linkage associated with the self-recognition traits. The genes within these regions could be filtered down to just two genes using genome scans for polymorphism and divergence.
Although Neurospora has favorable characteristics for linkage mapping studies, such as a small genome, the ability to cheaply maintain large immortal populations in lab, and haploid genetics, the difficulty in working with Neurospora—or any microbe—has been the identification of relevant phenotypes beyond simple traits. However, high-throughput sequencing has made it possible to use an unbiased “reverse ecology” approach to identifying adaptive phenotypes [75], in which genes with functions related to ecologically relevant traits are identified by examining genomic signatures of natural selection. An American N. crassa population bordering the Gulf of Mexico had been chosen for GWAS but when 50 of the isolates had been genotyped using RNAseq, analyses of population subdivision revealed multiple clusters and no cluster had sufficient sample size for a GWAS [23]. Fortunately, two clusters had at least 20 members, one in Louisiana and one further south in the Caribbean. Demographic inference based on a diffusion approximation to the site frequency spectrum [46] estimated a relatively higher population migration rate from Louisiana into the Caribbean (0.77 effective migrants per generation) than in the other direction, and a relatively recent divergence time (≈0.4 Mya) in agreement with the small proportion of fixed differences (9.4% of total SNPs; [23]). The Louisiana and Caribbean areas differ by 2–10° of latitude and winter temperatures are on average 9 °C cooler in Louisiana. The resulting hypothesis, that the Louisiana lineage had adapted to life at lower temperature, was not disproved by measuring fitness (i.e., growth) of isolates from each lineage at low (10 °C) and medium (25 °C) temperature. Genome scans to detect regions of extreme divergence in coding sequences, using measures of both relative and absolute divergence [76], revealed more than 30 such regions, but only 2 were identified by all divergence metrics. These two regions encompassed genes known to protect against cold temperatures: a cold shock RNA helicase and a prefoldin chaperone that, in yeast, protects actin from cold temperatures. To test the association between genotypes at candidate genes and fitness in cold temperature, the authors took advantage of the comprehensive N. crassa gene deletion collection to devise growth tests [77]. Among the eight genes found in the two regions of extreme genomic divergence, only the RNA helicase and prefoldin knockouts showed loss of cold tolerance.
The identification of genes underlying the ability of mycelia to distinguish self from nonself (i.e., allorecognition genes; see Note 6) in N. crassa by genomics and evolutionary approaches provides another example in which genes, and not phenotypes, were identified first. In filamentous fungi, allorecognition can result in somatic incompatibility (see Note 6), which is a type of programmed cell death that is triggered following fusion of genetically different cells. In N. crassa, somatic allorecognition controls both the fusion of germlings (as in the example of “germling regulated death” cited above) and the fusion of hyphae. Hyphal allorecognition is genetically controlled by the so-called het loci, and the het loci that have been characterized to date encode proteins carrying a HET domain (encoding a cell death effector) and show signatures of long-term balancing selection [78, 79]. Mining resequencing data from 26 Louisiana isolates for HET domain loci displaying elevated levels of variability, excess of intermediate frequency alleles, and deep gene genealogies identified 34 HET domain loci out of 69. Transformation, incompatibility assays, and genetic analyses revealed that one of the 34 candidates functioned as a het locus (het-e) that had been identified almost 50 years ago but awaited cloning [80]. The remaining 33 loci are, of course, prime targets for future investigations. These findings are encouraging and the collection of additional and more precise information on the biological and geographical origin of the samples, but also on relevant phenotypes (e.g., related to interactions with other microbes), should make it possible to exploit even better the potential of such reverse ecology approaches in the future.
6 Conclusion
Almost a century has passed since Shear and Dodge [9] described Neurospora, 70 years since Beadle and Tatum made it genetically conspicuous [14], and 50 years since Perkins et al. [17] initiated the global collections that fueled a host of research programs that continue to this day. More than 25 years ago, Perkins cautiously wrote that “Research on molecular, cellular and genetic mechanisms is certain to continue. It remains to be seen whether the promise of Neurospora for population genetics will be fulfilled.” Perkins would surely be happy to witness the tight integration between the two areas of research that now prevails, and to observe that the ways wild strains have proved useful for functional and evolutionary studies far exceeding what he and his collaborators anticipated when systematic collection was initiated in 1968. Much remains to be uncovered about the species richness of Neurospora and the distribution of known Neurospora species, although our understanding of Neurospora biogeography and speciation history is more advanced than it is for the vast majority of free-living microbes. The frequency of wild fires is not a limiting factor, providing a plethora of opportunities to collect new strains, and ongoing work also suggests that both conidial and aconidial species can be retrieved from soil. In the Perkins and Jacobson collection, the phenotype of F1 crosses to N. crassa is reported for more than 3600 isolates, and other phenotypes such as meiotic drive phenotype (i.e., spore killer type [66]), color, burned/nonburned substrate for more than 5000. Only a small fraction of this collection has been sequenced, and even with the many genomes already at hand, there is little indication of diminishing returns as additional species or populations are sequenced. The will be no lack of ecological or evolutionary questions. Most of the questions listed by Turner et al. [18] in their conclusion remain to be answered, and much remains to be done to dispel our ignorance about many aspects of the biology of microbial eukaryotes. We predict that, in the future, the message of hope of Perkins and Turner [4] “We are hopeful that the Neurospora work reviewed here will encourage wider studies in the genetics of fungal populations and will contribute to an increased appreciation of the potential contribution of the fungi.” will not need to be reiterated in any future review about the population genomics of Neurospora.
7 Notes
-
1.
An ascospore is a sexual spore of an ascomycete fungus, generated through meiosis (=meiospore). An ascus is a cell bearing ascospores. A perithecium is a spherical type of fruiting body in ascomycete fungi, containing ascus. A conidium is an asexual spore of a fungus, generated through mitosis (=mitospore).
-
2.
Mating type loci are genes that control sexual compatibility. Neurospora has two mating type alleles, referred to as Mat A et al.
-
3.
Homothallism defines situations where the successful fusion of gametes does not require functionally different mating-type alleles. Heterothallism defines situations where the successful fusion of gametes can occur only between haploids carrying functionally different mating-type alleles. Pseudohomothallism qualifies heterothallic species for which self-fertility is enforced by a modified program of meiosis that maintains a constant state of heterokaryosis, where nuclei of opposite mating type share a mycelium and are transmitted together in sexual or asexual spores.
-
4.
The reproductive system is the combination of the reproductive mode, the breeding system and the mating system [81]. The reproductive mode qualifies the process by which genes are transmitted across generation; reproductive mode can be asexual, sexual, or mixed when there is an alternation of sexual and asexual reproduction during the life cycle. The breeding system refers to the physiologic determinants of mating compatibility, regulated strictly, in fungi, in the haploid stage by mating-type loci. The breeding system of fungi can be heterothallic or homothallic, pseudohomothallism being a specific case of heterothallism (see Note 3). The mating system refers to the degree of genetic relatedness between mates. Outcrossing corresponds to the mating between cells derived from meioses in two different unrelated individuals, whereas inbreeding corresponds to the mating between related individuals. Inbreeding can be caused by selfing, the mating between meiotic products of the same diploid genotype, and several types of selfing can be distinguished in fungi [82]. Contrary to persistent misconceptions in the fungal literature, the breeding system has little influence on the mating system. For instance heterothallism, does not prevent selfing [27, 83].
-
5.
Heterokaryotic refers to multinucleate fungal cells that have two or more genetically different (but somatically compatible) nuclei. Homokaryotic refers to multinucleate fungal cells where all nuclei are genetically identical.
-
6.
Allorecognition refers to self–nonself recognition between conspecific individuals, while xenorecognition refers to self–nonself recognition between heterospecific individuals. Somatic incompatibility refers to the possible outcome of allorecognition processes, which limit successful somatic fusion to very closely related individuals or tissues [84].
References
Gladieux P, Ropars J, Badouin H, Branca A, Aguileta G, De Vienne DM et al (2014) Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol 23(4):753–773. https://doi.org/10.1111/Mec.12631
Jacobson DJ, Powell AJ, Dettman JR, Saenz GS, Barton MM, Hiltz MD et al (2004) Neurospora in temperate forests of western North America. Mycologia 96(1):66–74
Leducq JB (2014) Ecological genomics of adaptation and speciation in fungi. Adv Exp Med Biol 781:49
Perkins DD, Turner BC (1988) Neurospora from natural populations: toward the population biology of a haploid eukaryote. Exp Mycol 12(2):91–131
Taylor JW, Branco S, Gao C, Hann-Soden C, Montoya L, Sylvain I et al (2017) Sources of fungal genetic variation and associating it with phenotypic diversity. Microbiol Spectr 5(5)
Stajich JE, Berbee ML, Blackwell M, Hibbett DS, James TY, Spatafora JW et al (2009) The fungi. Curr Biol 19(18):R840–R8R5
Ad P (1843) Extrait d'un rapport adresse a M. Le Marechal Duc de Dalmatie, Ministre de la Guerre, President du Conseil, sur une alteration extraordinaire du pain de munition. Ann Chim Phys 9:5–21. 3rd Ser
Kuo H-C, Hui S, Choi J, Asiegbu FO, Valkonen JPT, Lee Y-H (2014) Secret lifestyles of Neurospora crassa. Sci Rep 4:5135. https://doi.org/10.1038/srep05135. http://www.nature.com/srep/2014/140530/srep05135/abs/srep05135.html-supplementary-information
Shear CL, Dodge BO (1927) Life histories and heterothallism of the red bread-mold fungi of the Monilia sitophila group. US Government Printing Office, Washington, DC
Davis RH (2007) Tending neurospora: David Perkins, 1919–2007, and Dorothy Newmeyer Perkins, 1922–2007. Genetics 175(4):1543–1548
Perkins DD, Davis RH (2002) Neurospora chronology 1843-2002. Fungal Genet Reports 49(1):4–8
Perkins DD (1992) Neurospora: the organism behind the molecular revolution. Genetics 130(4):687
Davis RH, Perkins DD (2002) Neurospora: a model of model microbes. Nat Rev Genet 3(5):397–403
Beadle GW, Tatum EL (1941) Genetic control of biochemical reactions in Neurospora. Proc Natl Acad Sci 27(11):499–506
Horowitz NH (1985) Roots: the origins of molecular genetics: one gene, one enzyme. BioEssays 3(1):37–39
Luque EM, Gutiérrez G, Navarro-Sampedro L, Olmedo M, Rodríguez-Romero J, Ruger-Herreros C et al (2012) A relationship between carotenoid accumulation and the distribution of species of the fungus Neurospora in Spain. PLoS One 7(3):e33658
Perkins DD, Turner BC, Barry EG (1976) Strains of neurospora collected from nature. Evolution 30(2):281–313. https://doi.org/10.2307/2407702
Turner BC, Perkins DD, Fairfield A (2001) Neurospora from natural populations: a global study. Fungal Genet Biol 32(2):67–92
Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D et al (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422(6934):859–868
Heller J, Zhao J, Rosenfield G, Kowbel DJ, Gladieux P, Glass NL (2016) Characterization of greenbeard genes involved in long-distance kind discrimination in a microbial eukaryote. PLoS Biol 14(4):e1002431
Palma-Guerrero J, Hall CR, Kowbel D, Welch J, Taylor JW, Brem RB et al (2013) Genome wide association identifies novel loci involved in fungal communication. PLoS Genet 9(8):e1003669
Zhao J, Gladieux P, Hutchison E, Bueche J, Hall C, Perraudeau F et al (2015) Identification of allorecognition loci in neurospora crassa by genomics and evolutionary approaches. Mol Biol Evol 32(9):2417–2432
Ellison CE, Hall C, Kowbel D, Welch J, Brem RB, Glass NL et al (2011) Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci 108(7):2831–2836
Jacobson DJ, Dettman JR, Adams RI, Boesl C, Sultana S, Roenneberg T et al (2006) New findings of Neurospora in Europe and comparisons of diversity in temperate climates on continental scales. Mycologia 98(4):550–559
Menkis A, Bastiaans E, Jacobson DJ, Johannesson H (2009) Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. J Evol Biol 22(9):1923–1936. https://doi.org/10.1111/j.1420-9101.2009.01801.x
Corcoran P, Dettman JR, Sun Y, Luque EM, Corrochano LM, Taylor JW et al (2014) A global multilocus analysis of the model fungus Neurospora reveals a single recent origin of a novel genetic system. Mol Phylogenet Evol 78:136–147. https://doi.org/10.1016/j.ympev.2014.05.007
Corcoran P, Jacobson DJ, Bidartondo MI, Hickey PC, Kerekes JF, Taylor JW et al (2012) Quantifying functional heterothallism in the pseudohomothallic ascomycete Neurospora tetrasperma. Fungal Biol 116:962–975. https://doi.org/10.1016/j.funbio.2012.06.006
Corcoran P, Anderson JL, Jacobson DJ, Sun Y, Ni P, Lascoux M et al (2016) Introgression maintains the genetic integrity of the mating-type determining chromosome of the fungus Neurospora tetrasperma. Genome Res 26(4):486–498
Dettman J, Jacobson D, Taylor J (2003) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57(12):2703–2720
Villalta CF, Jacobson DJ, Taylor JW (2009) Three new phylogenetic and biological Neurospora species: N. hispaniola, N. metzenbergii and N. perkinsii. Mycologia 101(6):777–789
Dettman JR, Jacobson DJ, Taylor JW (2006) Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia 98(3):436–446
Pringle A, Baker D, Platt J, Wares J, Latgé J, Taylor J (2005) Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59(9):1886–1899
Gladieux P, Feurtey A, Hood ME, Snirc A, Clavel J, Dutech C et al (2015) The population biology of fungal invasions. Mol Ecol 24(9):1969–1986. https://doi.org/10.1111/mec.13028
Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D (2006) Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos Trans R Soc Lond Ser B Biol Sci 361(1475):1947–1963. https://doi.org/10.1098/rstb.2006.1923
Taylor JW, Turner E, Pringle A, Dettman J, Johannesson H (2006) Fungal species: thoughts on their recognition, maintenance and selection. Fungi Environ:313–339
Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW (2003) Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57(12):2721–2741
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Nygren K, Strandberg R, Wallberg A, Nabholz B, Gustafsson T, García D et al (2011) A comprehensive phylogeny of Neurospora reveals a link between reproductive mode and molecular evolution in fungi. Mol Phylogenet Evol 59(3):649–663
Gladieux P, Wilson BA, Perraudeau F, Montoya LA, Kowbel D, Hann-Soden C et al (2015) Genomic sequencing reveals historical, demographic and selective factors associated with the diversification of the fire-associated fungus Neurospora discreta. Mol Ecol 24(22):5657–5675
Taylor JW, Fisher MC (2003) Fungal multilocus sequence typing—it’s not just for bacteria. Curr Opin Microbiol 6(4):351–356. https://doi.org/10.1016/s1369-5274(03)00088-2
Frichot E, Mathieu F, Trouillon T, Bouchard G, François O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196(4):973–983
Bryant D, Moulton V (2004) Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21(2):255–265. https://doi.org/10.1093/molbev/msh018
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4):1567–1587
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9(5):1322–1332. https://doi.org/10.1111/j.1755-0998.2009.02591.x
Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5(10):e1000695
Pandit A, Dubey PS, Mall S (2000) Sexual reproduction of yellow ecotype of Neurospora intermedia in nature. Fungal Genet Reports 47(1):81–82
Kitazima K (1925) On the fungus luxuriantry grown on the bark of the trees injured by the great fire of Tokyo on September 1, 1923. Japan J Phytopathol 1(6):15–19
Pandit A, Maheshwari R (1996) Life-history of Neurospora intermedia in a sugar cane field. J Biosci 21(1):57–79
Perkins DD (1994) Neurospora tetrasperma bibliography. Fungal Genet Reports 41(1):72–78
Perkins DD (2003) Neurospora tetrasperma bibliography—additions. Fungal Genet Reports 50(1):24–26
Dodge BO (1927) Nuclear phenomena associated with heterothallism and homothallism in the ascomycete Neurospora. J Agric Res 35:289–305
Glass NL, Metzenberg RL, Raju NB (1990) Homothallic Sordariaceae from nature: the absence of strains containing only theA mating type sequence. Exp Mycol 14(3):274–289
Dodge BO (1957) Rib formation in ascospores of Neurospora and questions of terminology. Bull Torrey Bot Club 84(3):182–188
Jacobson DJ (1995) Sexual dysfunction associated with outcrossing in Neurospora tetrasperma, a pseudohomothallic ascomycete. Mycologia:604–617
Raju NB (1992) Functional heterothallism resulting from homokaryotic conidia and ascospores in Neurospora tetrasperma. Mycol Res 96(2):103–116
Saenz GS, Stam JG, Jacobson DJ, Natvig DO (2001) Heteroallelism at the het-c locus contributes to sexual dysfunction in outcrossed strains of Neurospora tetrasperma. Fungal Genet Biol 34(2):123–129
Corcoran P (2013) Neurospora tetrasperma from natural populations: toward the population genomics of a model fungus. Acta Universitatis Upsaliensis, Uppsala
De Mita S, Siol M (2012) EggLib: processing, analysis and simulation tools for population genetics and genomics. BMC Genet 13(1):1
Akashi H, Osada N, Ohta T (2012) Weak selection and protein evolution. Genetics 192(1):15–31
Charlesworth D, Pannell J (2001) Mating systems and population genetic structure in the light of coalescent theory. In: Silvertown J, Westwood AJ (eds) Plants stand still but their genes don’t British ecological society special symposium 2000. Blackwell, Oxford, pp 73–96
Powell AJ, Jacobson DJ, Salter L, Natvig DO (2003) Variation among natural isolates of Neurospora on small spatial scales. Mycologia 95(5):809–819
Skupski MP, Jackson DA, Natvig DO (1997) Phylogenetic analysis of heterothallic Neurospora species. Fungal Genet Biol 21(1):153–162
Taylor JW, Natvig DO (1989) Mitochondrial DNA and evolution of heterothallic and pseudohomothallic Neurospora species. Mycol Res 93(3):257–272
Dodge BO (1928) Unisexual conidia from bisexual mycelia. Mycologia 20(4):226–234
Svedberg J (2017) Catching the spore killers: genomic conflict and genome evolution in neurospora, Digital comprehensive summaries of Uppsala Dissertations from the Faculty of Science and Technology. Acta Universitatis Upsaliensis, Uppsala, p 51
Jacobson DJ (2005) Blocked recombination along the mating-type chromosomes of Neurospora tetrasperma involves both structural heterozygosity and autosomal genes. Genetics 171(2):839–843
Swenson NG, Howard DJ (2005) Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat 166(5):581–591. https://doi.org/10.1086/491688
Roux C, Tsagkogeorga G, Bierne N, Galtier N (2013) Crossing the species barrier: genomic hotspots of introgression between two highly divergent Ciona intestinalis species. Mol Biol Evol 30(7):1574–1587
Turner E, Jacobson DJ, Taylor JW (2010) Reinforced postmating reproductive isolation barriers in Neurospora, an Ascomycete microfungus. J Evol Biol 23(8):1642–1656
Poggeler S, Masloff S, Jacobsen S, Kuck U (2000) Karyotype polymorphism correlates with intraspecific infertility in the homothallic ascomycete Sordaria macrospora. J Evol Biol 13(2):281–289
Turner E, Jacobson DJ, Taylor JW (2011) Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet 7(8):e1002204
Pomraning KR, Smith KM, Freitag M (2011) Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot Cell 10(6):724–733
Heller J, Clavé C, Gladieux P, Saupe SJ, Glass NL (2018) NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. PNAS 115(10):E2292–E2301
Li YF, Costello JC, Holloway AK, Hahn MW (2008) “Reverse ecology” and the power of population genomics. Evolution 62(12):2984–2994
Cruickshank TE, Hahn MW (2014) Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol 23(13):3133–3157
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L et al (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci 103(27):10352–10357
Glass NL, Dementhon K (2006) Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol 9(6):553–558
Muirhead CA, Glass NL, Slatkin M (2002) Multilocus self-recognition systems in fungi as a cause of trans-species polymorphism. Genetics 161(2):633–641
Wilson JF, Garnjobst L (1966) A new incompatibility locus in Neurospora crassa. Genetics 53(3):621–631
Neal PR, Anderson GJ (2005) Are ‘mating systems’ ‘breeding systems’ of inconsistent and confusing terminology in plant reproductive biology? Or is it the other way around? Plant Syst Evol 250(3-4):173–185. https://doi.org/10.1007/s00606-004-0229-9
Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T (2011) Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev 86(2):421–442
Giraud T, Yockteng R, Lopez-Villavicencio M, Refregier G, Hood ME (2008) Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryot Cell 7(5):765–775
Aanen DK, Debets AJM, de Visser J, Hoekstra RF (2008) The social evolution of somatic fusion. BioEssays 30(11-12):1193–1203
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this protocol
Cite this protocol
Gladieux, P., De Bellis, F., Hann-Soden, C., Svedberg, J., Johannesson, H., Taylor, J.W. (2020). Neurospora from Natural Populations: Population Genomics Insights into the Life History of a Model Microbial Eukaryote. In: Dutheil, J.Y. (eds) Statistical Population Genomics. Methods in Molecular Biology, vol 2090. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0199-0_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0199-0_13
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0198-3
Online ISBN: 978-1-0716-0199-0
eBook Packages: Springer Protocols