Abstract
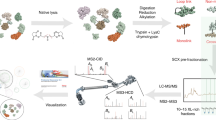
The detection and characterization of chemical adducts on proteins is of increasing interest. Here, we described a step-by-step procedure to identify unknown chemical adduct modifications on proteins resulting from the interaction with a given reactive compound. The protocol can be divided into two equally important parts: (1) the wet laboratory work, to produce high quality mass spectrometry (MS) data of in vitro modified proteins and (2) the dry laboratory work, to analyze the generated MS data and provide highly confident qualitative and quantitative results on the chemical composition and amino acid localization of adducts. This protocol is applicable to the study of any pharmaceutical or chemical compound forming covalent protein adducts, detectable in LC-MS/MS experiments.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Tornqvist M, Fred C, Haglund J et al (2002) Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J Chromatogr B Analyt Technol Biomed Life Sci 778:279–308
Acosta-Martin AE, Antinori P, Uppugunduri CR et al (2016) Detection of busulfan adducts on proteins. Rapid Commun Mass Spectrom 30:2517–2528
Hassan M, Andersson BS (2013) Role of pharmacogenetics in busulfan/cyclophosphamide conditioning therapy prior to hematopoietic stem cell transplantation. Pharmacogenomics 14:75–87
Huezo-Diaz P, Uppugunduri CR, Tyagi AK et al (2014) Pharmacogenetic aspects of drug metabolizing enzymes in busulfan based conditioning prior to allogenic hematopoietic stem cell transplantation in children. Curr Drug Metab 15:251–264
Tyanova S, Temu T, Cox J (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319
Acknowledgments
This study was supported by the Swiss Centre for Applied Human Toxicology and the Swiss National Science Foundation (grant No 32003B_143809). The authors would like to thank the Proteomics Core Facility of the University of Geneva for providing full access to the LTQ Orbitrap MS instrument.
None declared conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic Supplementary Material
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Antinori, P., Michelot, T., Lescuyer, P., Müller, M., Acosta-Martin, A.E. (2019). Detection of Unknown Chemical Adduct Modifications on Proteins: From Wet to Dry Laboratory. In: Evans, C., Wright, P., Noirel, J. (eds) Mass Spectrometry of Proteins. Methods in Molecular Biology, vol 1977. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-9232-4_8
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9232-4_8
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-9231-7
Online ISBN: 978-1-4939-9232-4
eBook Packages: Springer Protocols