Abstract
Introduction
High expression of aldehyde dehydrogenase1A1 (ALDH1A1) is observed in many organs and tumors and may identify benign and cancer stem cell populations.
Methods
In the current study, the stem cell characteristics were determined in cells isolated from human prostate cell lines and clinical prostate specimens based upon the ALDEFLUOR™ assay. Cells isolated based on the ALDEFLUOR™ assay were compared to cells isolated based on ATP binding cassette transporter G2 (ABCG2) activity using the side population assay. To test for stem cell characteristics of self-renewal and multipotency, cells with high and low ALDH1A1 activity, based on the ALDEFLUOR™ assay (ALDHHi and ALDHLow), were isolated from prostate clinical specimens and were recombined with rat urogenital sinus mesenchyme to induce prostate gland formation.
Results
The percentage of ALDHHi cells in prostate cell lines (RWPE-1, RWPE-2, CWR-R1, and DU-145) was 0.5 to 6%, similarly in non-tumor and tumor clinical specimens the percentage of ALDHHi cells was 0.6 to 4%. Recombinants using ALDHHi cells serially generated prostate tissue up to three generations with as few as 250 starting cells. Immunohistochemical analysis of the recombinants using ALDHHi cells contained prostatic glands frequently expressing androgen receptor (AR), p63, chromogranin A, ALDH1A1, ABCG2, and prostate specific antigen (PSA), compared to their ALDHLow counterparts. Inhibition of ALDH resulted in the reduction of sphere formation capabilities in the CWR-R1, but not in the RWPE-2 and DU-145, prostate cell lines. ABCG2 inhibition resulted in a more robust decrease of sphere formation in androgen sensitive cell lines, CWR-R1 and RWPE-2, but not androgen insensitive DU-145. ALDH1A1 expression was enriched in ALDHHi cells and non-side population cells. ABCG2 expression was only enriched in side population cells.
Conclusions
The percentage of ALDHHi cells in prostate cell lines and prostate tissue was consistently higher compared to cells with high ABCG2 activity, identified with the side population assay. The expression of the stem and differentiation markers indicates the ALDHHi recombinants contained cells with self-renewal and multipotency activity. When the two assays were directly compared, cells with the side population phenotype demonstrated more stem cell potential in the tissue recombination assay compared to ALDHHi cells. The increased stem cell potential of side population cells in the tissue recombination assay and the decrease in sphere formation when ABCG2 is inhibited indicates that the side population enriches for prostate stem cells.
Similar content being viewed by others
Introduction
Cytoprotective activity is proposed to protect stem cells from mutations that can arise from cytotoxic insults. Therefore, stem cells are often isolated based on cytoprotective activity. Conversely, stem cell assays have been developed exploiting a cytoprotective mechanism. Two examples of assays based on stem cell cytoprotective activity are: (1) the side population assay based on the efflux of Hoechst 33342 fluorescent dye by the ATP-binding cassette (ABC) transporters [1] and; (2) high activity of aldehyde dehydrogenases (ALDH) detoxifying enzymes [2]. The ALDEFLUOR™ assay was developed based on the second property [2]. ABC transporters remove cytotoxins and regulatory signals (Reviewed in [3]). ALDHs catalyze the irreversible oxidation of several different endogenously and exogenously produced aldehydes into the corresponding carboxylic acids (Reviewed in [4]).
The human ALDH superfamily consists of 19 members where ALDH1A1 and ALDH3A1 are thought to be important in stem cell protection, differentiation and expansion [2, 4–7]. Members of the ALDH family have been identified as markers for both normal and cancer stem cells in different tissues [4–7]. Specifically, ALDH1A1 and ALDH3A1 have been used as markers to isolate normal and cancer stem cells and have a potential functional role in normal and cancer stem cells (Reviewed in [4]). Van den Hoogen et al. determined expression of several ALDH enzyme isoforms in primary prostate tumors and primary prostate cultures and expression of ALDH1A1, ALDH4A1, ALDH7A1 and ALDH9A1 were elevated [8]. ALDH activity is emerging as a promising marker that can be used to identify and isolate normal and cancer stem cells [9].
The ALDEFLUOR™ assay is based on the conversion of Bodipy aminoacetaldehyde (BAAA), an ALDH substrate, to highly fluorescent Bodipy aminoacetate (BAA). Thus, cells with high ALDH1A1 activity (ALDHHi) can be isolated using fluorescence activated cell sorting (FACS) [2]. High levels of ALDH expression and activity in stem cells from various cancers including bladder, breast, lung, ovarian, liver, colon, pancreas and prostate have been detected using the ALDEFLUOR™ assay [10–18]. High ALDH1A1 activity is associated with poor prognosis in patients with breast and prostate cancer [18, 19]. Li et al. found that high ALDH1A1 expression in clinical prostate specimens correlated with poor prostate cancer patient survival and Gleason Score [18]. The studies also tested the tumorigenicity of ALDHHi and ALDHLow cells isolated from LNCaP and PC-3 prostate cancer cell lines. A minimum of 500 ALDHHi cells from LNCaP and PC-3 prostate cancer cell lines generated serially transplantable prostate tumors containing neuroendocrine and secretory cells, suggesting multipotentiality [18]. In separate studies, as few as 100 ALDHHi cells from the PC-3 M cell line had tumor forming and metastatic ability [8]. ALDHHi cells isolated from epithelial ovarian cancer cell lines are shown to have more sphere-forming capabilities, are more tumorigenic, and contribute to chemotherapy resistance as compared to ALDHLow cells. This suggests that ALDHHi cells are enriched for cancer stem-like cells [20]. The inhibition of ALDH1, with diethylaminobenzaldehyde (DEAB), reduced cancer stem cell characteristics of chemotherapy and radiation resistance in ALDHHi breast cancer cells [21].
In cell lines from different cancer types, ALDHHi cells have higher sphere formation ability, tumorigenicity and are more chemo- and radio-resistant compared to ALDHLow cells. However, ALDHHi cells have not been isolated from clinical specimens of prostate cancer, or tested in prostate stem cell assays. In the current study, we tested ALDHHi cells isolated from cell lines and human prostate specimens for stem cell properties. ALDHHi and ALDHLow cells from clinical specimens were evaluated for stem cell characteristics using tissue recombination with rat urogenital mesenchyme (rUGM) for serial prostate regenerative capability compared to cells isolated based on the side population assay. The requirement for ALDH and ABCG2 activity was determined in the sphere formation assay.
Methods
Ethics statement
All of the tissue samples were collected under an Institutional Review Board (IRB)-approved exempt non-human research protocol at Roswell Park Cancer Institute (RPCI). Specimens were collected after IRB-approved written consent from the patient was obtained at RPCI. All experiments were conducted and approved under our Institutional Animal Care and Use Committee (IACUC) at RPCI under protocol ID number 1201 M.
Cell lines and culture
Prostate cell lines RWPE-1 American Type Culture Collection (ATCC, Manassas, VA, USA), RWPE-2 (ATCC), CWR-R1 and DU-145 were maintained in culture medium as described previously [22]. Briefly, RWPE-1 and RWPE-2 cells were maintained in Keratinocyte Serum Free Medium supplemented with bovine pituitary extract and epidermal growth factor. DU-145 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 media supplemented with 10% FBS, L-glutamine, penicillin and streptomycin. CWR-R1 cells were maintained in Richter’s improved MEM supplemented with 2% FBS, HEPES, ITS, epidermal growth factor, linoleic acid and nicotinamide. All cell lines were cultured at 37°C in an atmosphere of 5% CO2. Cells were grown to 90% confluence and harvested by incubation with trypsin-EDTA.
Human prostate clinical specimen digestion
Fresh human benign prostate tissue and prostate cancer tissue (>90% cancer), identified by analysis of frozen sections from radical prostatectomy surgical specimens, was obtained from the Pathology Resource Network at RPCI as described previously [23]. All prostate tissue was obtained in accordance with National Institutes of Health (NIH) guidelines on the use of human subjects and with the approval of the RPCI Internal Research Board. Enzymatic tissue digestion with dispase and single cell suspension preparation was performed as described previously [22, 24].
Gating strategy used for FACS isolation of ALDHHi and ALDHLow cells
Cells isolated from prostate tissue were stained with the ALDEFLUOR™ reagent from the ALDEFLUOR™ assay kit (Stem Cell Technologies, Vancouver, BC, Canada) according to the manufacturer’s protocol and as previously described [2, 13]. The assay was optimized to interact with human ALDH1A1 according to the manufacturer’s protocol and as previously described [2, 13]. Briefly, the isolated cells were stained with 1 μM ALDEFLUOR™ reagent and incubated at 37°C for 45 to 60 minutes. RWPE-1 and RWPE-2 prostate cells were stained with 0.1 μM ALDEFLUOR™ reagent. CWR-R1 and DU-145 prostate cells were stained with 0.5 μM ALDEFLUOR™ reagent. After incubation, the cells were centrifuged at 950 × g and resuspended in 0.5 ml ALDEFLUOR™ assay buffer. The cells were labeled with 7-aminoactinomycin D (7-AAD) at a concentration of 5 μl/500 μl to eliminate dead cells. Cells were sorted into ALDHHi and ALDHLow populations with a BD FACS Aria II cell sorter (BD Bioscience, San Jose, CA, USA). Cells with high fluorescence indicate high ALDH1A1 activity, ALDHHi cells, were detected and the dead cells were eliminated using 7-AAD. DEAB (15 μM), a potent inhibitor of ALDH1 activity, was added to establish the ALDHHi population. Forward scatter and side scatter plots were used to reduce gating doublets and obtain a singlet population.
Gating strategy used for FACS isolation of side population and non-side population cells
Cells isolated from prostate tissue were stained with 10 μM Vybrant® DyeCycle™ Violet (DCV) (Invitrogen, Carlsbad, CA, USA) according to a protocol modified from Telford et al. [25] and 7-AAD as previously described [22, 26]. The gating strategy for sorting the side and non-side populations was performed as previously described [22, 26]. Viable cells were sorted based upon DCV content into side and non-side populations with a BD FACS Aria II cell sorter.
Sphere formation assay
The sphere formation assay was performed with RWPE-2, CWR-R1 and DU-145 prostate cells by treatment in the presence of 5, 10 and 20 μM DEAB or 1 and 5 μM Ko143. DEAB was prepared in 95% EtOH, and Ko143 was prepared in DMSO. ETOH (95% EtOH) or DMSO treatment was used as the vehicle control. Cells were plated at a density of 5,000 cells/well in ultra-low attachment 24-well plates (VWR-Cat # 29443–032, Radnor, PA, USA) in quadruplicate, and the experiment was performed two times. Cells were suspended in 40 μL medium and mixed thoroughly with 60 μL BD Matrigel™ (BD Bioscience, San Jose, CA, USA, Cat # 354234,). The mixture was pipetted around the rim of the well, and the plate was incubated in a 5% CO2 incubator at 37°C for 45 minutes to allow the BD Matrigel™ to solidify. A total of 900 μL of appropriate media was added to each well and plates were maintained in 5% CO2 incubator at 37°C for 10 to 14 days. Spheres in each well were quantitated after 10 to 14 days visually under a microscope at 10× magnification.
RNA isolation and cDNA synthesis
RNA isolated from the side population, non-side population, ALDHHi and ALDHLow cells CWR-R1 prostate cancer cells was used to determine the expression of ABCG2 and ALDH1 genes. RNA was isolated using RNAeasy micro kit (Qiagen, Valencia, CA, USA, Cat # 74004) according to the manufacturer’s protocol, and the concentration of total RNA was determined using the NanoDrop 8000 spectrophotometer (Wilmington, DE, USA). Total RNA (50 ng) from each sample was reverse transcribed into single-stranded cDNA with a SuperScript III First-Strand cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA, Cat, # 18080–051) using oligo (dT) primers, according to the manufacturer’s protocol.
Real-time PCR
Real-time PCR was performed to determine gene expression. The synthesized cDNA was amplified by real-time PCR using a SYBR Green master mix kit (Applied Biosystems, Carlsbad, CA, USA, Cat # 4309155). The reactions were carried out in a total volume of 20 μL. Analyses for ABCG2, ALDH1A1, ALDH4A1, ALDH7A1, ALDH9A1 and GAPDH were performed. Conditions for the PCR were: 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles each run at 95°C for 15 seconds and 60°C for 1 minute. The primers (Integrated DNA Technologies, Coralville, IA, USA) used were: ABCG2 (0.25 μM) sense, 5′ – AGCAGGATAAGCCACTCATAGA – 3′;
ABCG2 (0.25 μM) antisense, 5′ – GTTGGTCGTCAGGAAGAAGAG – 3′;
ALDH1A1 (0.5 μM) sense, 5′ – TTACCTGTCCTACTCACCGATT – 3′;
ALDH1A1 (0.5 μM) antisense, 5′ – GCCTTGTCAACATCCTCCTTAT – 3′;
ALDH4A1 (0.5 μM) sense, 5′ – GGACGTGCAGTACCAAGTGT – 3′;
ALDH4A1 (0.5 μM) antisense, 5′ – TAGGCTTCAGGTCCCACTCT – 3′;
ALDH7A1 (0.5 μM) sense, 5′ – CAACGAGCCAATAGCAAGAG – 3′;
ALDH7A1 (0.5 μM) antisense, 5′ – GCATCGCCAATCTGTCTTAC – 3′;
ALDH9A1 (0.5 μM) sense, 5′ – GCTGCCAGGATAATAAGGGA – 3′;
ALDH9A1 (0.5 μM) antisense, 5′ – GAAATGTCAATGTCCAAGCG – 3′;
GAPDH (0.2 μM) sense, 5′ – GAACATCATCCCTGCCTCTACT – 3′;
GAPDH (0.2 μM) antisense, 5′ – CGCCTGCTTCACCACCTT -3′.
Reactions were run and analyzed using an Applied Biosystems 7300 fast real-time PCR system (Carlsbad, CA, USA). Gene expression was normalized to GAPDH and fold change in gene expression was calculated using the following formula: fold change in gene expression = 2-(ΔΔdCt).
Tissue recombinant technique
Rat urogenital sinus mesenchyme was dissected as described previously [27, 28]. First generation recombinants were made by aliquoting 50 to 2,000 ALDHHi or ALDHLow cells or 50 to 250 side population or non-side population cells isolated from human prostate tissue specimens with a single rUGM in a 200 μL microcentrifuge tube. Sorted cells were spun onto the rUGM, or in a separate tube sorted cells were pelleted alone as a negative control. Sorted cells + rUGM and sorted cell pellets were suspended in collagen as described previously [26].
Renal grafting
Cells plus rUGM or cells alone in collagen were grafted under the renal capsule in 12-week-old SCID male CbySmn. CB 17 Prkdcscid mice, which were castrated and implanted subcutaneously with 12.5 mg sustained-release testosterone pellets (Innovative Research of America, Sarasota, FL, USA) as described previously [26]. Following 6 to 12 weeks of growth, grafts were harvested and weighed. When ductal growth was present, grafts were micro-dissected and a portion was used for serial recombination with new rUGM as described previously [26]. Briefly, an approximately 1 mm cube of tissue containing both epithelial and stromal compartments was micro-dissected from the graft and used for serial recombination with new rUGM [26]. The remaining portion of the recombinant was fixed in neutral buffered formalin as described previously [26].
Identification of the origin of prostate generated using fluorescence in situ hybridization (FISH)
In order to identify the cells contributing to the prostate epithelium in the tissue recombinants FISH was performed (Additional file 1: Figure S1). Mammalian telomeres were detected as described previously [26]. The number of human prostatic glands in each recombinant was determined as previously described [26].
Immunohistochemistry (IHC)
Immunohistochemical analysis was performed to examine the expression of prostate differentiation markers as described previously [26]. ALDH1A1 was detected using a 1:80 dilution of rabbit monoclonal anti-ALDH1A1 (cat # NB110-55451, Novus Biologicals, Littleton, CO, USA). Human prostate tissue served as a positive control, and no primary antibody controls served as negative controls in all experiments (Additional file 2: Figure S2).
Aperio imaging recombinant histology and IHC
All slides containing stained recombinants were scanned on the ScanScope XT System in the Pathology Resource Network (Buffalo, NY, USA) at RPCI, and images were analyzed with Spectrum Version 10.2.2.2317 (Vista, CA, USA).
Statistical analysis
Fisher’s exact test, Student’s t-test and One-way ANOVA were performed with GraphPad Prism version 6.00 software (La Jolla, CA, USA). The recombinant survival rate was defined as the proportion of recombinants demonstrating growth at each generation (cycle) analyzed. Recombinants with growth in the third generation were censored. The Kaplan-Meier analysis performed using SAS v9.3 software (Cary, NC, USA) provides visual comparison of these survival distributions for the ALDHHi and ALDHLow recombinants. The null hypothesis of no difference in the survival distributions was assessed using the Logrank test.
Results
Isolation of ALDHHi and ALDHLow cells from prostate cell lines
The ALDEFLUOR™ assay was performed on RWPE-1 RWPE-2, CWR-R1 and DU-145 prostate cell lines. ALDHHi cells represented 0.5 to 6% of the total cell population in RWPE-1, RWPE-2, CWR-R1 and DU-145 prostate cells (Figures 1A, C, E, G). ALDHHi cells are gated based upon the absence of the population in the presence of DEAB, an inhibitor of ALDH1, (Figures 1B, D, F, H) and the ALDHLow cells were gated with a clear separation from the ALDHHi cells (Figures 1A, C, E, G).
Isolation of ALDHHiand ALDHLowcells from prostate cells by ALDEFLUOR™ assay. RWPE-1, RWPE-2, CWR-R1 and DU-145 prostate cells were stained with ALDEFLUOR™ reagent and analyzed using BD FACS Aria II sorter. ALDHHi cells were detected in the higher fluorescent region (A, C, E and G). ALDHHi population was inhibited with DEAB resulting in no ALDHHi cell detection (B, D, F and H). The ALDHLow population is gated with a clear separation from the ALDHHi population.
Prostate generation in recombinants with ALDHHi and ALDHLow cells isolated from human prostate specimens
The ALDEFLUOR™ assay was performed on six radical prostatectomy specimens. ALDHHi cells represented 0.6 to 4% of the cells in non-tumor, tumor and mixed prostate specimens (Figure 2 and Table 1). ALDHHi or ALDHLow cells were recombined with rUGMs in the tissue recombinant assay in order to functionally test for stem cell properties. Sorted cells suspended in collagen without rUGM were used as a non-inductive control to test tumorigenic capacity. Equal numbers of cells (50 to 2,000) were used for each sorted population (Table 2). At the end of each grafting period, the grafts were harvested, weighed and examined microscopically, blinded to the contributing population, for glandular formation. Recombinants that demonstrated ductal growth were micro-dissected, and a small portion (approximately 1 mm3) of the ductal structure was recombined with new rUGM and implanted under the renal capsule of host mice. The remaining portion of each recombinant was formalin-fixed and paraffin-embedded for IHC analysis. If under the dissecting microscope there was no evidence of ductal growth or the ductal growth was obviously rodent, the recombinant was processed for histological analysis of microscopic ductal growth, and no serial recombination with rUGM was performed. This procedure resulted in some recombinants with histological growth not being serially passed and some recombinants being serially passed, although there was no histological evidence of human epithelial cell growth.
Isolation of ALDHHiand ALDHLowcells from clinical human prostate specimen by ALDEFLUOR™ assay. Cells isolated from clinical human prostate specimen were stained with ALDEFLUOR™ reagent and analyzed using BD FACS Aria II sorter. ALDHHi cells were detected in the higher fluorescent region (A, C and E). ALDHHi population was inhibited with DEAB resulting in no ALDHHi cell detection (B, D and F). ALDHLow population is gated with a clear separation from the ALDHHi population.
Survival rate and number of human prostate glands generated by recombinants
All recombinants were stained with H&E to evaluate prostatic gland structure. Rodent telomere repeats were used to identify rodent epithelial cell contribution to the prostatic glands in the recombinants. Each recombinant was examined for expression of rodent telomere repeat with FISH analysis (Additional file 1: Figure S1). Prostate glands that were negative for rodent telomeres were considered to be of human origin [29]. First generation growth was analyzed in 83 recombinants, in the first generation 14/42 (approximately 33%) ALDHHi recombinants and 10/41 (approximately 24%) ALDHLow recombinants contained human epithelial ductal growth, but the difference was not statistically significant (Table 2, Figure 3A). All recombinant growth was evaluated by a genitourinary pathologist (GA) and was considered benign. There was no evidence of prostate cancer pathology in any of the specimens regardless of the pathology of the initial tissue used. All recombinants were stained with alpha-methylacyl-CoA racemace (AMACR), a marker of prostate cancer and prostatic intraepithelial neoplasia and no recombinants had detectable expression of AMACR (data not shown). The total number of human prostatic glands in each first generation recombinant was determined (Figure 3B), but there was no significant difference in the average number of human prostatic glands in recombinants generated with ALDHHi cells compared to ALDHLow cells.
Number of human prostate glands generated per recombinant and recombinant survival rate of recombinants grafted. A. The percentage of recombinants with human ductal growth in the first generation ALDHHi n = 42 and ALDHLow n = 41, analyzed by Fisher’s Exact test P = 0.4692. B. The number of glands per recombinant in the first generation derived from ALDHHi cells (n = 6) and ALDHLow (n = 5) was quantitated and analyzed by unpaired t-test analysis P = 0.2558. C. Kaplan-Meier plot of recombinant survival rate in multiple generations, Logrank test P = 0.3890.
The number of ALDHLow sorted cells used in tissue recombination or the length of engraftment did not seem to impact the frequency of ductal growth in the recombinants (Table 3). However, when 500 to 2,000 ALDHHi cells were used in recombination, the ductal growth rate increased to 50 to 60% compared to 0% to 19% when 50 to 250 ALDHHi cells were used (Table 3). Recombinants with a minimum of 250 ALDHHi cells serially generated prostate up to three generations, whereas no recombinant with ALDHLow cells generated prostate ductal growth in the third generation (Table 3). The frequency of recombinant growth in the second and third generations was calculated as a percentage of the initial number of recombinants grafted in the first generation (Table 3).
The ability of the ALDHHi and ALDHLow cells to generate prostatic tissue in serial generations was compared using Kaplan-Meier methods and was not statistically significant P = 0.389 (Figure 3C). When recombinants were harvested there was no definitive way to determine the species of origin of the ductal growth in the recombinants. Therefore, serial passage of recombinants was only performed when non-rodent ductal growth was evident upon micro-dissection. This resulted in instances when recombinants that demonstrated histological human epithelial growth were not recombined and other occasions where the ductal growth that was serially passaged did not contain human epithelium upon histological and FISH analysis. These recombinants were removed from analysis if human ductal growth was not detected in serial generations. If human ductal growth was detected in the second and/or third generation recombinants, human ductal growth was assumed in previous generations even if not histologically detected in the remaining portion not used for serial recombination.
ALDHHi and ALDHLow recombinants grafted under the renal capsule of SCID mice generated differentiated human prostate glands
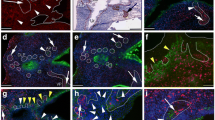
All recombinants with human prostatic growth were analyzed for prostate differentiation markers. Prostate differentiation was tested by examining expression of p63, AR, PSA, ALDH1A1, ABCG2 and chromogranin A. Most glands contained cells expressing p63 and AR (Figure 4B, C). Recombinants generated from ALDHHi cells were more likely to contain cells also expressing ALDH1A1, PSA, ABCG2 or chromogranin A (Table 4). A recombinant generated with 2,000 ALDHHi cells from 3 NT (Table 1) had cells positive for all markers examined (Figure 4). High ALDH1A1 expression was observed in few recombinants in isolated cells outside the p63 expressing basal layer (Figure 4F).
IHC analysis of prostate differentiation markers in a recombinant using 2,000 ALDHHicells. IHC analysis of prostate differentiation markers in a recombinant using 2,000 ALDHHi cells: A. H & E staining; B. p63 IHC; C. AR IHC; D. PSA IHC; E. ALDH1A1 IHC; F. Higher magnification of boxed area of ALDH1A1 IHC in E; G. ABCG2 IHC; H. Higher magnification of boxed area of ABCG2 IHC in G; I. Chromogranin A (Chr A) IHC; J. Higher magnification of Chromogranin A IHC in a different region of same recombinant. Arrow indicates positive cell. Scale bar = 50 μm.
Nineteen percent of the ALDHHi recombinants contained ALDH1A1 expressing cells within prostatic glandular structure compared to only approximately 2% of ALDHLow recombinants (Table 4). Thus, results from tissue recombinant experiments showed that ALDHHi cells serially generated prostate tissue, and ALDH1A1 expressing cells are retained in the prostatic growth providing evidence of self-renewal.
Inhibition of ALDH and ABCG2 activity decreases the sphere-forming capability of prostate cancer cells
To determine the requirement of ALDH function in stem cell maintenance, sphere formation capabilities were tested in the presence and absence of DEAB, an ALDH1 inhibitor. Sphere formation capability of RWPE-2, CWR-R1 and DU-145 prostate cells was detected upon inhibition of ALDH by DEAB. RWPE-1 cells did not form spheres (data not shown). Though sphere formation was significantly decreased in the CWR-R1 cells upon inhibition of ALDH activity (Figure 5C), sphere formation was not decreased in either RWPE-2 (Figure 5A) or DU-145 (Figure 5E) prostate cells. RWPE-2 had the highest percentage of ALDHHi cells (Figure 1C) suggesting ALDH activity may not be important in stem cell function.
Sphere formation assay. RWPE-2, CWR-R1, and DU-145 prostate cells were treated in the presence of DEAB at 5, 10 and 20 μM or Ko143 at 1 and 5 μM concentration or vehicle control. Spheres were counted in each well after 10 to 14 days of growth. Sphere forming ability of RWPE-2 cells did not decrease upon treatment with DEAB (A). Sphere formation was decreased in RWPE-2 cells upon treatment with Ko143 at 5 μM concentration (B). Sphere formation was decreased in CWR-R1 cells upon treatment with DEAB (C) or Ko143 (D). Sphere forming ability of DU-145 cells did not decrease upon treatment with DEAB (E) or Ko143 (F).
The side population assay is another functional assay used to isolate stem cells. For comparison, ABCG2 function was inhibited in the prostate cell lines with Ko143. Sphere formation was significantly decreased in both RWPE-2 (Figure 5B) and CWR-R1 cells (Figure 5D) upon inhibition of ABCG2 activity. Sphere formation was not altered in DU-145 cells (Figure 5F) indicating AR may be required for ABCG2 regulation of stem cell maintenance. Taken together ALDH activity represents a marker of prostate stem cells but ALDH activity may not be required for stem cell maintenance.
Gene expression in cells isolated by stem cell functional assays
Real-time PCR was performed on CWR-R1 cells isolated by the ALDEFLUOR™ and side population assays to determine the expression of ABCG2 and specific ALDH isoforms. The CWR-R1 cell line was chosen because sphere formation depended on both ALDH and ABCG2 activity. ABCG2 expression was approximately five-fold higher in the side population compared to the non-side population, thus the side population assay selects for cells with high ABCG2 expression (Figure 6A). ALDH1A1 gene expression was approximately 30-fold higher in ALDHHi cells compared to ALDHLow cells indicating the ALDEFLUOR™ assay enriches for cells with high ALDH1A1 expression (Figure 6B). The expression of ALDH4A1, ALDH7A1 and ALDH9A1 genes was not significantly altered either the side or ALDH populations (Figure 6A, B). Additionally, ALDH1A1 gene expression was higher in non-side population cells compared to the side population (Figure 6A). Thus the ALDEFLUOR™ assay does not select for cells expressing ABCG2 and side population assay does not select for cells with expressing ALDH1A1.
Gene expression in ALDHHi, ALDHLow, side population, and non-side population cells isolated from CWR-R1 cells. ABCG2, ALDH1A1, ALDH4A1, ALDH7A1 and ALDH9A1 gene expression was detected by real-time PCR in CWR-R1 cells isolated by the side population and ALDEFLUOR™ assays. Gene expression was normalized to GAPDH and fold change in gene expression in each population was calculated based on the formula: 2-(ΔΔdCt). ABCG2 expression was the highest in the side population with 5.4-fold increase compared to non-side population (A), whereas ALDH1A1 expression was highest in NSP (A) and ALDHHi cells (B). Approximately 26-fold and 30-fold increase in ALDH1A1 gene expression was observed in NSP (A) and ALDHHi cells (B) respectively as compared to their counter populations. Expression of ALDH4A1, ALDH7A1 and ALDH9A1 genes was not significantly different in any of the populations (A and B). NSP, Non-Side Population; SP, Side Population.
Comparison of cells isolated based upon ALDEFLUOR™ and side population assays
To compare stem cell properties of cells sorted based upon the ALDEFLUOR™ or the side population assays, cells were isolated from the same clinical specimens using either assays. Single viable cells from two radical prostatectomy specimens were analyzed with ALDEFLUOR™ and side population assays. The ALDHHi and ALDHLow cells and side and non-side population cells were isolated from specimens 5NT and 6 T by FACS (Figure 7 and Table 1) and used in tissue recombination.
Isolation of ALDHHi, ALDHLow, side population and non-side population cells from clinical human prostate specimen. Cells isolated from clinical human prostate specimen were stained with either DCV or ALDEFLUOR™ reagent. The ALDHHi cells were detected in the higher fluorescent region (A). ALDHHi population was inhibited with DEAB resulting in no ALDHHi cell detection (B). Side population was gated based upon its absence in the presence of FTC, a specific inhibitor of ABCG2 (C and D).
Recombinants with as low as 50 side population cells could serially generate human prostate tissue for two generations (Table 5). Recombinants with a minimum of 250 ALDHHi cells isolated from the same clinical specimen were required to generate human prostate tissue when serially recombined for two generations (Table 5). Third generation growth was not detected in either population (data not shown). The difference in the efficiencies of prostatic ductal growth in recombinants with ALDHHi and side population cells is intriguing but needs to be further investigated to determine if one assay is more efficient at enriching stem cells compared to the other.
Discussion
In the current study, we have analyzed the stem cell properties of cells with high ALDH activity with the ALDEFLUOR™ assay and the role of ALDH in stem cell maintenance. Additionally, we have compared the stem cell properties of cells isolated with the ALDEFLUOR™ assay to cells isolated based upon ABCG2 activity, with the side population assay. The requirement for ALDH and ABCG2 activity for stem cell properties was evaluated in tumorigenic prostate cell lines by determining sphere forming capability when ALDH is inhibited by DEAB or ABCG2 is inhibited by Ko143.
As few as 250 ALDHHi cells from freshly digested and sorted clinical human prostate specimens recombined with rUGM generated prostate tissue for three generations. The serial prostate regenerative capability of ALDHHi cells increased when higher numbers (500 to 2,000 ALDHHi cells) were used in tissue recombination assays. Similarly, studies using breast and prostate cancer cell lines showed low tumor growth frequency using 500 ALDHHi cells, and tumor growth frequency was increased when higher numbers (5 × 104 to 1 × 105) of ALDHHi cells were used [8, 18, 30, 31]. Furthermore, only tumors derived from ALDHHi PC-3 cells are composed of both ALDHHi and ALDHLow cells demonstrating differentiation capability [18]. The percentage of ALDHHi cells obtained in our studies in RWPE-1, RWPE-2, CWR-R1 and DU-145 prostate cell lines ranged from 0.5 to 6%. These ranges are consistent with previous studies with PC-3 and LNCaP prostate cell lines [18]. We identified side populations in RWPE-2 (0.4%) (Additional file 3: Figure S3), and previously we have identified the side population in RWPE-1 (0.04%), CWR-R1 (0.4%) and DU-145 (0.73%) cell lines [22]. The percentage of cells in the side population is lower in most cell lines, except in the androgen insensitive DU-145 cell line. Thus, the side population appears to be more selective compared to the ALDEFLUOR™ assay in prostate cell lines.
Real-time PCR analysis indicates that ABCG2 expressing cells are not enriched in the ALDHHi population, and ALDH1A1 expression is not enriched in the side population (Figure 6A, B). Thus, the cells isolated with the two assays were different populations. Both functional assays were not performed simultaneously because the assays are not compatible [2]. This is in contrast to the hematopoietic system, wherein quiescent CD34+CD38Low side population cells were highest in the ALDHHi population, and the combination of assays produces the most enriched primitive hematopoietic stem/progenitor cell population [32].
The sphere formation assay has been shown as an in vitro correlate to test for the presence of stem cells [33]. ALDHHi cells from ovarian, breast and prostate cell lines have more sphere-forming and tumorigenicity capability compared to ALDHLow cells [18–20, 30, 31, 33, 34]. Sphere formation decreased in CWR-R1 cells upon treatment with DEAB, suggesting that ALDH activity is important for maintaining stem cell properties in CWR-R1 prostate cells. Sphere-forming ability of RWPE-2 and DU-145 prostate cells did not decrease upon treatment with DEAB. Concurrently, we tested the requirement of ABCG2 activity for sphere formation using Ko143, an ABCG2 specific inhibitor. ABCG2 activity is required for the formation of spheres in RWPE-2 and CWR-R1 prostate cells. However, the sphere-forming ability of androgen insensitive DU-145 prostate cells did not decrease upon treatment with Ko143, suggesting the requirement of androgen signaling for ABCG2 dependent stem cell properties. ABCG2 mediated androgen efflux regulates nuclear AR expression [35]. Thus, ABCG2 activity in AR deficient cells may not be required for stem cell properties.
Recombinants with ALDHHi cells had human ductal growth in the first generation at a high frequency, and each recombinant contained a high number of glands compared to the recombinants generated with ALDHLow cells. Results from IHC analysis showed that ALDHHi recombinants contained ALDH1A1 expressing cells, thus demonstrating self-renewal capabilities, nuclear AR and the basal cell marker p63. Additionally, ALDH1A1 expression was also detected in non-basal cells in the recombinants as illustrated by cytoplasmic staining. An ALDHHi recombinant showed that ALDH1A1 positive glands also expressed ABCG2, PSA and chromogranin A. These results suggest that ALDH1A1 is expressed in heterogeneous, cells with multipotency, some of which possess stem cells properties. As further evidence of differentiation potential, a higher percent of ALDHHi recombinants with human ductal growth expressed ALDH1A1, ABCG2 and PSA with 19%, 45% and 33% recombinants showing expression, respectively, compared to 2%, 29% and 15%, respectively, in ALDHLow recombinants (Table 4).
None of the recombinants suspended in collagen alone grew prostatic tissue and recombinants with rUGM did not demonstrate prostate cancer pathology. This is not surprising since the presence of an inductive microenvironment, provided by the rUGM in these experiments, has been shown to be critical to trigger prostate generation [36, 37]. Previous studies have shown that the presence of inductive mesenchyme can normalize transformed epithelial cells giving rise to normal prostatic growth even from cells isolated from tumor specimens [38–40]. Alternatively, prostate cancer pathology was observed in the tissue recombination assay with rUGM when a high number of prostate cancer cells were used [37]. There could be several possibilities for why no cancer pathology was found in the recombinants using cells isolated based on either the ALDEFLUOR™ or side population assays. Several studies used prostate cancer cell lines, increasing the possibility of isolating cancer stem cells. The number of cells used in this study, 50 to 2,000, may not be enough to implant the rare cancer stem cell. There could also be patient variability with respect to the presence of cancer stem cell markers accounting for the presence of fewer side populations or ALDHHi cells in different specimens. Alternatively, there could be cancer stem cells expressing different markers, other than ABCG2 or ALDH activity, that have the capability to initiate prostate tumor growth. Prostate cancer stem cells from human specimens capable of forming prostate tumor pathology in the recombination assay were not isolated by the side population or ALDEFLUOR™ assays. Nevertheless, results from the tissue recombination assay showed that there is serial transplantable and prostate regenerative capability in recombinants with side population and ALDHHi cells.
Previous reports from our laboratory have identified side population cells in different prostate cell lines and freshly digested human prostate specimens [22, 26]. Furthermore, side population cells isolated from human prostate specimens generated prostatic tissue for multiple generations in the tissue recombination assay [26]. Therefore, we wanted to directly compare putative prostate stem cells isolated based upon ABCG2 or ALDH activity. Since, the ALDH product formed in the ALDEFLUOR™ reaction is effluxed by the ABCG2 transporter [2], the side population and the ALDEFLUOR™ assays were performed separately. The specimens contained 1% ALDHHi cells and about 0.3% of the cells in the specimens demonstrated the side population phenotype. Results from the tissue recombination assay with side population cells are consistent with our previous studies [26]. In our current study, we have observed serial generation of prostate growth with as low as 250 ALDHHi and 50 side population cells. While recombinants with as low as 50 side population cells generated prostate for two generations, a minimum of 250 ALDHHi cells were required to serially generate prostate for two generations. Results from the tissue recombinant assay comparing the prostate regeneration capability of side population and ALDHHi cells suggested that a higher percentage of cells with ABCG2 activity have stem cell potential compared to cells with high ALDH activity.
Conclusions
This is the first study to demonstrate that ALDHHi cells isolated from human prostate tissue demonstrate prostate tissue growth in the tissue recombination assay. Cells isolated based on ABCG2 activity have a higher frequency of stem cell potential compared to ALDHHi cells and the ALDEFLUOR™ assay does not enrich for ABCG2 expressing cells. The ALDEFLUOR™ assay does isolate prostate stem cells, but the side population assay enriches for prostate cells with more stem cell potential.
Abbreviations
- 7-AAD:
-
7-Amino actinomycin D
- ABCG2:
-
ATP binding cassette transporter G2
- ALDH:
-
Aldehyde dehydrogenase
- ALDH1A1:
-
Aldehyde dehydrogenase1A1
- ALDHHi:
-
High aldehyde dehydrogenase1A1 activity isolated based upon ALDEFLUOR™ assay
- ALDHLow:
-
Low aldehyde dehydrogenase1A1 activity isolated based upon ALDEFLUOR™ assay
- AMACR:
-
Alpha-MethylAcyl-CoA Racemace
- AR:
-
Androgen receptor
- DCV:
-
Dye cycle violet
- DEAB:
-
Diethyl amino benzaldehyde
- FACS:
-
Fluorescence activated cell sorting
- FISH:
-
Fluorescence in situ hybridization
- IHC:
-
Immunohistochemistry
- PCR:
-
Polymerase chain reaction
- PSA:
-
Prostate specific antigen
- RPCI:
-
Roswell park cancer institute
- rUGM:
-
Rat urogenital mesenchyme.
References
Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC: Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996, 183: 1797-1806. 10.1084/jem.183.4.1797.
Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C: Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA. 1999, 96: 9118-9123. 10.1073/pnas.96.16.9118.
Dean M: ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia. 2009, 14: 3-9. 10.1007/s10911-009-9109-9.
Moreb JS: Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther. 2008, 3: 237-246. 10.2174/157488808786734006.
Muramoto GG, Russell JL, Safi R, Salter AB, Himburg HA, Daher P, Meadows SK, Doan P, Storms RW, Chao NJ, McDonnell DP, Chute JP: Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010, 28: 523-534.
Douville J, Beaulieu R, Balicki D: ALDH1 as a functional marker of cancer stem and progenitor cells. Stem Cells Dev. 2009, 18: 17-25. 10.1089/scd.2008.0055.
Sladek NE: Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003, 17: 7-23. 10.1002/jbt.10057.
van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, van der Pluijm G: High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010, 70: 5163-5173. 10.1158/0008-5472.CAN-09-3806.
Corti S, Locatelli F, Papadimitriou D, Donadoni C, Salani S, Del Bo R, Strazzer S, Bresolin N, Comi GP: Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006, 24: 975-985. 10.1634/stemcells.2005-0217.
Chang B, Liu G, Xue F, Rosen DG, Xiao L, Wang X, Liu J: ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009, 22: 817-823.
Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G: ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007, 1: 555-567. 10.1016/j.stem.2007.08.014.
Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM: Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009, 69: 3382-3389. 10.1158/0008-5472.CAN-08-4418.
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, Katz RL: Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009, 7: 330-338. 10.1158/1541-7786.MCR-08-0393.
Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, Hong SM, Koorstra JB, Rajeshkumar NV, He X, Goggins M, Iacobuzio-Donahue C, Berman DM, Laheru D, Jimeno A, Hidalgo M, Maitra A, Matsui W: Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010, 102: 340-351. 10.1093/jnci/djp535.
Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z, Stass SA, Jiang F: Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010, 19: 327-337. 10.1158/1055-9965.EPI-09-0865.
Lingala S, Cui YY, Chen X, Ruebner BH, Qian XF, Zern MA, Wu J: Immunohistochemical staining of cancer stem cell markers in hepatocellular carcinoma. Exp Mol Pathol. 2010, 89: 27-35. 10.1016/j.yexmp.2010.05.005.
Burger PE, Gupta R, Xiong X, Ontiveros CS, Salm SN, Moscatelli D, Wilson EL: High aldehyde dehydrogenase activity: a novel functional marker of murine prostate stem/progenitor cells. Stem Cells. 2009, 27: 2220-2228. 10.1002/stem.135.
Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, Stass SA, Jiang F: ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2010, 90: 234-244. 10.1038/labinvest.2009.127.
Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL: High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2008, 13: 2236-2252.
Wang YC, Yo YT, Lee HY, Liao YP, Chao TK, Su PH, Lai HC: ALDH1-bright epithelial ovarian cancer cells are associated with cd44 expression, drug resistance, and poor clinical outcome. Am J Pathol. 2012, 180: 1159-1169. 10.1016/j.ajpath.2011.11.015.
Croker AK, Allan AL: Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res Treat. 2011, 133: 75-87.
Mathew G, Timm EA, Sotomayor P, Godoy A, Montecinos VP, Smith GJ, Huss WJ: ABCG2-mediated DyeCycle Violet efflux defined side population in benign and malignant prostate. Cell Cycle. 2009, 8: 1053-1061. 10.4161/cc.8.7.8043.
Morrison C, Cheney R, Johnson CS, Smith G, Mohler JL: Central quadrant procurement of radical prostatectomy specimens. Prostate. 2009, 69: 770-773. 10.1002/pros.20925.
Gangavarpu KJ, Huss WJ: Isolation and applications of prostate side population cells based on dye cycle violet efflux. Curr Protoc Toxicol. 2011, Chapter 22: Unit 22 22-
Telford WG, Bradford J, Godfrey W, Robey RW, Bates SE: Side population analysis using a violet-excited cell-permeable DNA binding dye. Stem cells. 2007, 25: 1029-1036. 10.1634/stemcells.2006-0567.
Foster BA, Gangavarapu KJ, Mathew G, Azabdaftari G, Morrison CD, Miller A, Huss WJ: Human prostate side population cells demonstrate stem cell properties in recombination with urogenital sinus mesenchyme. PLoS One. 2013, 8: e55062-10.1371/journal.pone.0055062.
Foster BA, Donjacour AA: Techniques for Modeling Human Cancer in Mice: a Practical Guide for Dissections, Microsurgery and Other Biomethods. DVD compilation of techniques from “Techniques for Modeling Human Cancer in Mice”: workshop held at Jackson Laboratory. 2002, Mouse Models of Human Cancers Consortium, NCI: Bar Harbor, Maine
Staack A, Donjacour AA, Brody J, Cunha GR, Carroll P: Mouse urogenital development: a practical approach. Differentiation. 2003, 71: 402-413. 10.1046/j.1432-0436.2003.7107004.x.
Vander Griend DJ, Konishi Y, De Marzo AM, Isaacs JT, Meeker AK: Dual-label centromere and telomere FISH identifies human, rat, and mouse cell contribution to multispecies recombinant urogenital sinus xenografts. Prostate. 2009, 69: 1557-1564. 10.1002/pros.21001.
Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS: Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010, 16: 45-55. 10.1158/1078-0432.CCR-09-1630.
Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS: Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69: 1302-1313. 10.1158/0008-5472.CAN-08-2741.
Pierre-Louis O, Clay D, Brunet De La Grange P, Blazsek I, Desterke C, Guerton B, Blondeau C, Malfuson JV, Prat M, Bennaceur-Griscelli A, Lataillade JJ, Le Bousse-Kerdiles MC: Dual SP/ALDH functionalities refine the human hematopoietic Lin-CD34 + CD38- stem/progenitor cell compartment. Stem Cells. 2009, 27: 2552-2562. 10.1002/stem.186.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS: In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17: 1253-1270. 10.1101/gad.1061803.
Garraway IP, Sun W, Tran CP, Perner S, Zhang B, Goldstein AS, Hahm SA, Haider M, Head CS, Reiter RE, Rubin MA, Witte ON: Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010, 70: 491-501.
Huss WJ, Gray DR, Greenberg NM, Mohler JL, Smith GJ: Breast cancer resistance protein-mediated efflux of androgen in putative benign and malignant prostate stem cells. Cancer Res. 2005, 65: 6640-6650. 10.1158/0008-5472.CAN-04-2548.
Hayward SW, Haughney PC, Rosen MA, Greulich KM, Weier HU, Dahiya R, Cunha GR: Interactions between adult human prostatic epithelium and rat urogenital sinus mesenchyme in a tissue recombination model. Differentiation. 1998, 63: 131-140. 10.1046/j.1432-0436.1998.6330131.x.
Toivanen R, Berman DM, Wang H, Pedersen J, Frydenberg M, Meeker AK, Ellem SJ, Risbridger GP, Taylor RA: Brief report: a bioassay to identify primary human prostate cancer repopulating cells. Stem Cells. 2011, 29: 1310-1314. 10.1002/stem.668.
Wong YC, Cunha GR, Hayashi N: Effects of mesenchyme of the embryonic urogenital sinus and neonatal seminal vesicle on the cytodifferentiation of the Dunning tumor: ultrastructural study. Acta Anat. 1992, 143: 139-150. 10.1159/000147240.
Hayashi N, Cunha GR, Wong YC: Influence of male genital tract mesenchymes on differentiation of Dunning prostatic adenocarcinoma. Cancer Res. 1990, 50: 4747-4754.
Hayashi N, Cunha GR: Mesenchyme-induced changes in the neoplastic characteristics of the Dunning prostatic adenocarcinoma. Cancer Res. 1991, 51: 4924-4930.
Acknowledgements
This work was supported by NYSTEM (CO24292) to WJH; NIH (R01CA095367) to BAF; and NCI Cancer Center Support Grant (CA016056) to RPCI. We thank RPCI Pathology Resource Network for clinical specimens; RPCI Mouse Tumor Model Resource for assistance with tissue processing, sphere formation and animal studies; RPCI Flow and Image Cytometry Core for FACS; Earl Timm for flow cytometry and FACS; Ellen Karasik for assistance in recombination technique and histological preparations; Bryan Gillard for animal care; and Dr. James Mohler for the CWR-R1 cells.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors have read the journal’s policy and have the following conflicts: WJH is a RPCI patent holder for “Methods for evaluating and implementing prostate disease treatments” Patent No. 8048640. All other authors do not have any conflicting interests.
Authors’ contributions
KJG, BAF and WJH conceived and designed the experiments. KJG, BAF and WJH performed the experiments and analyzed the results. GA and CDM performed pathology analysis. AM performed statistical analysis. CDM contributed the reagents/materials/analysis tools. KJG, BAF and WJH wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
13287_2013_480_MOESM1_ESM.tiff
Additional file 1: Figure S1: Positive and negative controls for telomere FISH analysis. Mouse control tissue A. Telomere FISH analysis positive for telomere repeats; B. DAPI counter stain same field; Human control tissue C. Telomere FISH analysis negative for telomere repeats; D. DAPI counter stain same field. Scale bar = 10 μm. (TIFF 2 MB)
13287_2013_480_MOESM2_ESM.tiff
Additional file 2: Figure S2: Positive and negative human prostate controls for IHC staining. A. p63 IHC; B. no primary antibody (p63) with goat anti-mouse IgG antibody IHC; C. AR IHC; D. no primary antibody (AR) with goat anti-rabbit IgG antibody IHC; E. PSA IHC; F. no primary antibody (PSA) with goat anti-mouse IgG antibody IHC; G. ABCG2 IHC; H. no primary antibody (ABCG2) with goat anti-mouse IgG antibody IHC; I. Chromogranin A IHC; J. no primary antibody (Chromogranin A) with goat anti-rabbit IgG antibody IHC. K. ALDH1A1 IHC; L. no primary antibody (ALDH1A1) with goat-anti-rabbit IgG antibody IHC. Scale bar = 50 μm. (TIFF 4 MB)
13287_2013_480_MOESM3_ESM.tiff
Additional file 3: Figure S3: Isolation of side population and non-side population cells from RWPE-2 prostate cells. RWPE-2 prostate cells were stained with DCV reagent. The side population was gated based upon its absence in the presence of FTC, a specific inhibitor of ABCG2 (A and B). (TIFF 207 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Gangavarapu, K.J., Azabdaftari, G., Morrison, C.D. et al. Aldehyde dehydrogenase and ATP binding cassette transporter G2 (ABCG2) functional assays isolate different populations of prostate stem cells where ABCG2 function selects for cells with increased stem cell activity. Stem Cell Res Ther 4, 132 (2013). https://doi.org/10.1186/scrt343
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/scrt343