Abstract
Following reports of an invasive snail causing crop damage in the expansive Mwea irrigation scheme in Kenya, samples of snails and associated egg masses were collected and sent to CABI laboratories in the UK for molecular identification. DNA barcoding analyses using the cytochrome oxidase subunit I gene gave preliminary identification of the snails as Pomacea canaliculata, widely considered to have the potential to be one of the most invasive invertebrates of waterways and irrigation systems worldwide and which is already causing issues throughout much of south-east Asia. To the best of our knowledge, this is the first documented record of P. canaliculata in Kenya, and the first confirmed record of an established population in continental Africa. This timely identification shows the benefit of molecular identification and the need for robust species identifications: even a curated sequence database such as that provided by the Barcoding of Life Data system may require additional checks on the veracity of the underlying identifications. We found that the egg mass tested gave an identical barcode sequence to the adult snails, allowing identifications to be made more rapidly. Part of the nuclear elongation factor 1 alpha gene was sequenced to confirm that the snail was P. canaliculata and not a P. canaliculata/P. maculata interspecies hybrid. Given the impact of this species in Asia, there is need for an assessment of the risk to Africa, and the implementation of an appropriate response in Kenya and elsewhere to manage this new threat to agriculture and the environment.
Similar content being viewed by others
Background
Apple snails (Ampullariidae) are freshwater gastropods distributed naturally throughout the humid tropics and subtropics (Berthold 1991). Pomacea, the largest of nine extant genera in the family, are New World species native to South, Central and North America (Hayes et al. 2015). Some Pomacea species, mainly from South America, demonstrate notable invasiveness outside of their native ranges (Hayes et al. 2008; Wu et al. 2010; Lv et al. 2013; Yang et al. 2018). Of these, Pomacea canaliculata (Lamarck, 1822), is listed among ‘100 of the world’s worst invasive species’ (Lowe et al. 2000) and together with the morphologically similar P. maculata Perry, 1810 (P. insularum (d’Orbigny, 1835) is a junior synonym), have become serious agricultural and ecological pests, causing significant economic losses in wetland rice cultivation and threatening biodiversity (Cowie 2002; Tamburi and Martín 2009; Qiu et al. 2011). Historically, descriptions of Pomacea have been based on the conchological characteristics of a few specimens, resulting in taxonomic confusion (Cowie et al. 2006). Furthermore, identification and differentiation between species has proved extremely difficult (Thiengo et al. 1993; Cazzaniga 2002; Cowie et al. 2006) since morphological similarities are shared among many different species and they also exhibit substantial intraspecific and geographic variation (Hayes et al. 2012; Mahilum and Demayo 2014). In light of this, phylogenetic analyses have provided an invaluable framework for discovering and delineating apple snail species (Rawlings et al. 2007; Hayes et al. 2008; Yang and Yu 2019). By correctly identifying species, pest and natural resource managers are provided with a better scientific foundation on which to implement effective management plans to mitigate spread, and minimize their agricultural and environmental impacts. Apple snails continue to spread to new areas and have been reported recently in Europe (López et al. 2010; EFSA 2020) and Ecuador (Horgan et al. 2014), but the African continent appeared to remain hitherto free of established populations (Seuffert and Martin 2017). Cowie et al. (2017) cited Wu and Xie (2006) as an unconfirmed report of P. canaliculata’s presence in Egypt, and they also cited Berthold (1991) reporting its presence in South Africa, noting that it was identified as P. lineata but was probably P. canaliculata, and that it was possibly not established. Similarly, they noted that there had been unconfirmed reports of Marisa cornuarietis being found in Sudan and Tanzania (Cowie et al. 2017).
Methods
The aim of the study was to determine the identity of the snails found at two locations (Tebere and Ndekia) in Mwea irrigation scheme in Kenya and to determine if they were indigenous or a potentially invasive species.
Sample collection and handling
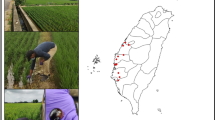
Field surveys for apple snails were carried out in Kenya in September 2020 after reports that an unknown snail species had invaded close to 222 ha of the adjacent Tebere and Ndekia sections of the 10,117 ha Mwea irrigation scheme in Kirinyaga County. Sampling sites included irrigated rice fields and other host plants at the Scheme. At each sampling location, latitude, longitude and altitude were determined using a global positioning system device (Garmin, USA) (Fig. 1; Table 1). At each sample collection site, photographs were taken of the adults (e.g. Fig. 2a) and eggs (e.g. Fig. 2c). Samples of eggs and adults were also collected for morphological and molecular identification (Figs. 2b, d, e). In total, 39 samples were collected from the two affected areas (Table 1). Preliminary morphological identification to genus level was done using features described by Hayes et al. (2012). Egg masses were collected from non-aquatic plants, partially submerged posts and aerial plant projections (e.g. Fig. 2c), as these snails lay their eggs out of water. Collection was undertaken carefully thereby avoiding early crushing/breaking of the eggs. A sample of the collected snails was rinsed in running tap water to remove soil and other debris, then transferred to clean containers and immersed in absolute ethanol (> 99% v/v). The lids of the containers were tightened and the containers were stored in a laboratory refrigerator (at 2–8 °C). Egg masses were also collected and treated in the same way.
Of the samples collected, 15 adult snails and two egg masses were prepared for dispatch to the UK (with associated documentation allowing export for identification of the specimens from KEPHIS and CABI’s UK (Defra) licence for importing material from overseas) whilst representative specimens were deposited in the National Museums of Kenya (NMK), Nairobi, Kenya, under the voucher specimen number NMK/INV/MOL-012090. During packing, the ethanol was drained and the samples placed on dry cotton wool in a plastic container for shipment to CABI’s diagnostic and molecular biology laboratories in the UK.
On receipt in the UK, samples were processed as follows:
Specimens were unpacked, subjected to visual examination and photographed. Three adult snails, broadly covering the range of the sizes seen but otherwise selected at random, were selected for identification as was one sample of a partial egg mass. Snail bodies were removed carefully from the shells, using alcohol-sterilised, then flamed, forceps and immersed in absolute ethanol (> 99% v/v) in individual sterile tubes. The selected egg mass was immersed in absolute ethanol (> 99% v/v) in a separate tube. All tubes were transferred to a laboratory freezer (− 20 °C) in which they were stored for 48 h, to ensure no living material remained.
On retrieval from the freezer, several small sections of each snail sample were removed from clean areas (foot and tentacles, not the digestive system), using alcohol-sterilised fine forceps and scalpel, taking care to avoid the digestive system and other organs. One egg was removed for processing. These sub-samples were placed in fresh sterile 1.5 ml microcentrifuge tubes containing absolute ethanol (> 99% v/v) and placed in the freezer until they could be processed further.
Molecular methodology
Samples were removed from the tubes and gently blotted dry on clean lint-free tissue paper. The samples were then subjected to DNA extraction using the DNeasy Blood and Tissue kit (Qiagen, UK) following the manufacturer’s protocol, amended to incorporate an overnight incubation at 55 °C once the buffer ATL and proteinase K had been added to the tissue (Polaszek et al. 2014). Subsequent steps were as prescribed by the manufacturer.
PCR reactions were carried out using a Hybaid PCR Express thermal cycler in heated-lid mode. Amplifications were carried out in 0.5 ml microcentrifuge tubes in 20 µl reactions containing: 1 µl DNA extract; Primers LCO1490 and HCO2198 (5′-GGTCAACAAATCATAAAGATATTGG-3′ and 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′, respectively; Folmer et al. 1994) each at 150 nM; and 10 μl of MegaMix-Royal (Microzone Ltd, UK) mastermix solution, containing an optimised mixture of Taq polymerase in 2 × Enhancing Buffer (6 mM MgCl2), with 400 μM dNTPs and blue MiZN loading dye. Reactions were made up to a final volume of 20 μl with sterile molecular grade H2O. PCR reactions were preincubated for 5 min at 95 °C followed by 39 cycles of: 30 s at 94 °C; 30 s at 51 °C and 75 s at 72 °C. Samples were finally incubated for 10 min at 72 °C, then cooled and finally held at 10 °C for 1 h before PCR products were visualised with gel electrophoresis, purified by microCLEAN (Microzone Ltd., UK), and resuspended in 15 µl sterile molecular grade H2O. After sequencing reactions, excess unincorporated dye terminators were removed by means of DyeEx 2.0 (Qiagen, UK) gel filtration columns, according to the manufacturer’s instructions. Eluted samples were resuspended in 16 µl highly deionised formamide. Sanger sequencing was undertaken using an ABI 3130 Genetic Analyser in accordance with the manufacturer’s instructions. Sequence trace files were examined for quality using the DNA Sequencing Analysis Software 6 (Applied Biosystems, UK), then exported as text files.
Data analysis
Sequences obtained in the present study were screened against the holdings of NCBI-GenBank and EMBL-EBI using the BLAST (Altschul et al. 1997) and FASTA (Pearson 1990) algorithms, respectively. Sequences were then compared with representative voucher specimen sequences obtained from the Barcoding of Life Data system (BOLD; http://www.boldsystems.org/; Ratnasingham and Hebert 2007) and additional sequences from the GenBank data base (http://www.ncbi.nlm.nih.gov/genbank/). Sequences were aligned using the multiple sequence alignment tools CLUSTALW (Thompson et al. 1994) and MUSCLE (Edgar 2004) in MEGA7 (Kumar et al. 2016) and these were then optimized manually in the MEGA7 program. For the COI sequences, phylogenetic relationships were inferred by means of the maximum likelihood method based on the Tamura–Nei model (Tamura and Nei 1993). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor–Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, before selecting the topology with superior log likelihood value. Codon positions included were first, second and third regions, whilst all positions containing gaps and missing data were eliminated. Trees obtained were drawn to scale, with branch lengths measured in the number of substitutions per site. The bootstrap consensus tree was inferred from 1000 replicates and is taken to represent the evolutionary history of the taxa analysed (Felsenstein 1985). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the respective branches (Felsenstein 1985). Further evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016). The EF1a sequences were subjected to phylogenetic analysis and bootstrap analysis in the same way as for the COI sequences. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 16 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 395 positions in the final dataset.
Results
The four samples processed for sequencing (three adult snails plus one egg from an egg mass) gave identical sequences when barcoded with the COI primers. When screened against the holdings of GenBank and BOLD, all top matches, showing > 99% identity to the Mwea samples, were to sequences assigned to P. canaliculata with the sole exception of what appeared to be an aberrant P. maculata sequence (MK992483) from Uruguay (data not shown). This sequence showed in the GenBank/BLAST and BOLD results but did not feature in the EMBL-EBI/FASTA top 500 results. This sequence (as well as further instances of apparently confounding results within the respective Barcode Index Number system (BINs) of species within this genus), suggests that even a curated database such as BOLD may harbour some misidentified or incorrectly assigned sequences (e.g. Pentinsaari et al. 2020). The MK992483 sequence also appeared confounding in the phylogenetic analysis undertaken subsequently where it was placed within the P. canaliculata cluster and not with the rest of P. maculata, (including sequences labelled as P. insularum, which is now considered a junior objective synonym of P. maculata (CABI 2021)). It should be noted, however, that broad acceptance of the utility of DNA barcodes for distinguishing between species is not the same as recommending it as replacement for robust polyphasic taxonomic research: a database is only as good as the information on which it is built. Hayes (2021) has recently stated the case clearly: that DNA-based tools can have great benefits in terms of objectivity and cost-effective processing when used correctly but, if used inappropriately, they can lead to greater confusion that may hinder efforts to e.g. stop the spread of an invasive pest or pathogen.
The phylogenetic analyses enabled inferences to be made regarding the relationships among the sequences screened and the placement of the Kenyan sequences within the broader genus arrangement. The tree with the highest log likelihood (− 6485.26) is shown as Fig. 3. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The analysis involved 71 nucleotide sequences. All positions containing gaps and missing data were eliminated. In total, there were 546 positions in the final dataset.
The EF1a analysis was undertaken in a similar way. This analysis involved 13 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 395 positions in the final dataset. The tree with the highest log likelihood (− 1030.82) is shown as Fig. 4.
These results show clearly that the samples tested are P. canaliculata. Sequences have been deposited in GenBank with the accession numbers MW363565-MW363568 (COI) and MW589392 (EF1a).
Discussion
Early detection of non-native invasive species is a critical factor to inform timely implementation of contingency and control measures and allow the greatest chance of successful management or eradication. The gradual erosion in morpho-taxonomic expertise worldwide, as well as the potential for confusion caused by taxa that look almost identical, such as the species of Pomacea (Estebenet et al. 2006), can make positive identification a challenge (Rama Rao et al. 2018). Molecular approaches have contributed to improved taxonomic understanding and clarification of the distribution and invasion pathways of Pomacea species (Kannan et al. 2020) and provide an invaluable and complementary diagnostic tool for the timely identification of these highly destructive invasive species. Concerns have been raised about the quality of some sequences in the global, open access sequence databases such as EMBL, GenBank and DDBJ and the relevant assigned identifications (e.g. Bridge et al. 2003). The aforementioned databases have been improving their acceptance criteria for sequences of late but, even so, BOLD has been considered to be more authoritative for many taxa as it involves greater curation of the submitted sequences. Recent studies on the quality of identifications from GenBank and BOLD noted potential issues with both (Meiklejohn et al. 2019; Pentinsaari et al. 2020). It is likely that this misidentification accounts for the phylogenetic placement of the P. maculata sequence (MK992483) described above, within the P. canaliculata cluster. These results reinforce the need for polyphasic approaches to taxonomy, combining molecular and morphometric analyses to ensure robust species descriptions. This, in turn, requires that global sequence repositories are built on solid criteria which can then lead to identification methodologies that are accurate and reliable.
This appears to be the first confirmed report of P. canaliculata in continental Africa, although it is present in La Réunion and Mauritius (EPPO 2020).
The arrival of this notorious invasive species in mainland Africa raises many questions that cannot be dealt with in detail in this short report. One is how might it have arrived in Kenya? In Asia it is thought to have been illegally but intentionally introduced to Taiwan around 1980, with subsequent rapid spread to other countries in South-East Asia “predominantly human-mediated” (Cowie 2002), often because it was seen as a potential food source. Unconfirmed media reports in Kenya suggest that the snail was introduced to control weeds, but no permit to import the species has been issued by the Kenya Standing Technical Committee on Imports and Exports.
In South-East Asia, snails have caused significant crop losses in rice, resulting in annual management costs of more than US$1200 million (Brito and Joshi 2016). In Kenya farmers are already complaining about the damage caused to rice in Mwea, where over 70% of the country’s rice is grown (Atera et al. 2018). In the initial area of infestation, farmers have reported up to 92% damage to newly transplanted rice seedlings. Experience in Asia indicates that damage decreases as seedling age increases and is higher where direct seeding is used rather than transplanting seedlings (CABI 2020). Pomacea canaliculata also displays developmental and reproductive plasticity allowing adaptation to a wide range of habitats with different climates and the magnitude of its environmental and agricultural damage is climatically linked and predicted to increase globally (Lei et al. 2017).
How far might it have already spread in Kenya? Delimiting surveys are in progress, but the Mwea rice production area drains into the Tana River, a designated Ramsar wetland site and an important ecosystem in East Africa in which P. canaliculata could have significant impacts. Although renowned for the damage it causes to agricultural production, there are many actual and potential environmental impacts of P. canaliculata (Carlsson 2017; Martin et al. 2019) including direct effects through herbivory, competition and predation, but also indirect effects such as transmission of human disease and bioaccumulation of pollutants.
Having reached mainland Africa, there must also be concern that the snail will be transported to other countries in the continent, including in West Africa where nearly two-thirds of Africa’s rice is produced (Sers and Mughal 2020). Rice consumption in Africa is increasing, and although the gap has been reduced, the continent is still unable to produce sufficient rice to meet demand (Senthilkumar et al. 2020). No environmental suitability modelling for P. canaliculata in Africa has been conducted, although models have been constructed elsewhere that could be used to examine the potential spread (Gilioli et al. 2017). A risk assessment for P. canaliculata in Africa is needed as a priority for which such modelling would be a useful input.
In the meantime, plans for containment measures and management should be a priority in the sections/area already infested, in order to prevent spread. Further, the National Plant Protection Organisations in Africa should be on the alert for this species. Based on a risk assessment, phytosanitary measures should be implemented to reduce the likelihood of its introduction, but at the same time contingency plans should be prepared so that it can be detected and responded to early and effectively should it reach other countries.
Conclusions
The results obtained from our barcoding analysis show clearly that P. canaliculata is present in Kenya. Given this snail’s ongoing reputation as one of the top 100 invasive species threats worldwide (Lowe et al. 2000), such an early finding has real significance for plant health in Kenya requiring urgent remedial action.
We have also clearly demonstrated the benefits of DNA barcoding for detecting and verifying such organisms in the absence of expert morphometric malacologists due to the benefits of the global sequence repositories such as BOLD. The matching results obtained from the egg showed that a barcoding approach enables significant time savings as there is no need to grow the egg to maturity for definitive identification.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BLAST:
-
Basic local alignment search tool
- BOLD:
-
Barcoding of life datasystems
- COI:
-
Cytochrome oxidase c subunit I gene
- DDBJ:
-
DNA database of Japan
- dNTPs:
-
Deoxyribonucleotide triphosphates
- EBI:
-
European Bioinformatics Institute
- EF1a:
-
Elongation factor 1-alpha gene
- EMBL:
-
European Molecular Biology Laboratory
- ML:
-
Maximum likelihood
- NCBI:
-
National Center for Biotechnology Information
- PCR:
-
Polymerase chain reaction
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Atera EA, Onyancha FN, Majiwa EBO. Production and marketing of rice in Kenya: challenges and opportunities. J Dev Agric Econ. 2018;10:64–70.
Berthold T. Vergleichende Anatomie, Phylogenie und historische Biogeographie der Ampullariidae (Mollusca, Gastropoda). Abh Nat Ver (NF). 1991;29:1–256.
Bridge PD, Roberts PJ, Spooner BM, Panchal G. On the unreliability of published DNA sequences. New Phytol. 2003;160:43–8. https://doi.org/10.1046/j.1469-8137.2003.00861.x.
Brito FC, Joshi RC. The golden apple snail Pomacea canaliculata: a review on invasion, dispersion and control. Outlooks Pest Manag. 2016;27(4):157–63.
CABI. Pomacea canaliculata [original text by J Litsinger (1996), reviewed by R Joshi (2003) and R Cowie (2013)]. In: Invasive species compendium. Wallingford, UK: CAB International. 2020. www.cabi.org/isc. Accessed 11 Dec 2020.
CABI. Pomacea maculata [original text by RL Burks, AE Miller, AL Hill (2013), reviewed by R Cowie (2015)]. In: Invasive Species Compendium. Wallingford, UK: CAB International. 2021. www.cabi.org/isc. Accessed 08 Feb 2021.
Carlsson NOL. lnvasive apple snails are threatening natural ecosystems in Southeast Asia, Europe and North America. In: Joshi RC, Cowie RH, Sebastian LS, editors. Biology and management of invasive apple snails. Muñoz, Philippines: Philippine Rice Research Institute; 2017. p. 45–62.
Cazzaniga NJ. Old species and new concepts in the taxonomy of Pomacea (Gastropoda: Ampullariidae). Biocell. 2002;26:71–81.
Cowie RH. Apple snails (Ampullariidae) as agricultural pests: their biology, impacts and management. In: Barker GM, editor. Molluscs as crop pests. Wallingford, UK; CABI Publishing; 2002. pp. 145–192. http://www.cabi.org/cabebooks/ebook/20023046840. https://doi.org/10.1079/9780851993201.0145.
Cowie RH, Hayes KA, Thiengo SC. What are apple snails? Confused taxonomy and some preliminary resolution. In: Joshi RC, Sebastian LS, editors. Global advances in ecology and management of golden apple snails. Munoz, Nueva Ecija: PhilRice; 2006. p. 3–23.
Cowie RH, Hayes KA, Strong EE, Thiengo SC. Non-native apple snails: systematics, distribution, invasion history and reasons for introduction. In: Joshi RC, Cowie RH, Sebastian LS, editors. Biology and management of invasive apple snails. Muñoz, Philippines: Philippine Rice Research Institute; 2017. p. 3–32.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
EFSA (European Food Safety Authority), Schrader G, Delbianco A and Vos S. Pest survey card on Pomacea spp. EFSA supporting publication 2020:EN-1877. 37 pp. https://doi.org/10.2903/sp.efsa.2020.EN-1877.
EPPO, 2020. EPPO Global database. In: EPPO Global database, Paris, France: EPPO. https://gd.eppo.int/taxon/POMACA/distribution. Accessed on 18 Dec 2020.
Estebenet AL, Martín PR, Burela S. Conchological variation in Pomacea canaliculata and other South American Ampullariidae (Caenogastropoda, Architaenioglossa). Biocell. 2006;30:329–35.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Gilioli G, Pasquali S, Martín PR, et al. A temperature-dependent physiologically based model for the invasive apple snail Pomacea canaliculata. Int J Biometeorol. 2017;61:1899–911. https://doi.org/10.1007/s00484-017-1376-3.
Hayes KA. Taxonomic shortcuts lead to long delays in species discovery, delineation, and identification. Biol Invasions. 2021. https://doi.org/10.1007/s10530-020-02438-8.
Hayes KA, Joshi RC, Thiengo SC, Cowie RH. Out of South America: multiple origins of non-native apple snails in Asia. Divers Distrib. 2008;14(4):701–12. https://doi.org/10.1111/j.1472-4642.2008.00483.
Hayes KA, Cowie RH, Thiengo SC, Strong EE. Comparing apples with apples: clarifying the identities of two highly invasive neotropical Ampullariidae (Caenogastropoda). Zool J Linn Soc-Lond. 2012;166(4):723–53. https://doi.org/10.1111/j.1096-3642.2012.00867.
Hayes KA, Burks RL, Castro-Vazquez A, Darby PC, Heras H, Martín PR, et al. Insights from an integrated view of the biology of apple snails (Caenogastropoda: Ampullariidae). Malacologia. 2015;58:245–302. https://doi.org/10.4002/040.058.0209.
Horgan FG, Felix MI, Portalanza DE, Sanchez L, Rios WMM, Farah SE, Wither JA, Andrade CI, Espin EB. Responses by farmers to the apple snail invasion of Ecuador’s rice fields and attitudes toward predatory snail kites. Crop Prot. 2014;62:135–43.
Kannan A, Rama Rao S, Ratnayeke S, Yow YY. The efficiency of universal mitochondrial DNA barcodes for species discrimination of Pomacea canaliculata and Pomacea maculata. Peer J. 2020;8:e8755. https://doi.org/10.7717/peerj.8755.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Lei J, Chen L, Li H. Using ensemble forecasting to examine how climate change promotes worldwide invasion of the golden apple snail (Pomacea canaliculata). Environ Monit Assess. 2017;189:404.
López MA, Altaba CR, Andree KB, López V. First invasion of the apple snail Pomacea insularum in Europe. Tentacle. 2010;18:26–8.
Lowe S, Browne M, Boudjelas S, De Poorter M. 100 of the World’s worst invasive alien species a selection from the global invasive species database. The Invasive Species Specialist Group of the Species Survival Commission of the World Conservation Union. 2000;12 pp.
Lv S, Zhang Y, Liu HX, Hu L, Liu Q, Wei FR, Guo YH, Steinmann P, Hu W, Zhou XN, Utzinger J. Phylogenetic evidence for multiple and secondary introductions of invasive snails: Pomacea species in the People’s Republic of China. Divers Distrib. 2013;19:147–56. https://doi.org/10.1111/j.1472-4642.2012.00924.x.
Mahilum JJM, Demayo CG. Sexual dimorphism on shell shape of Pomacea canaliculata Lamarck thriving in lakes using the geometric morphometric approach. Int J Biosci Biochem Bioinforma. 2014;4(4):284–9.
Martín PR, Burela S, Seuffert ME, Tamburi NE, Saveanu L. Invasive Pomacea snails: actual and potential environmental impacts and their underlying mechanisms. CAB Rev. 2019;14(042):1–11.
Meiklejohn KA, Damaso N, Robertson JM. Assessment of BOLD and GenBank—their accuracy and reliability for the identification of biological materials. PLoS ONE. 2019;14:e0217084. https://doi.org/10.1371/journal.pone.0217084.
Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Method Enzymol. 1990;183:63–98. https://doi.org/10.1016/0076-6879(90)83007-V.
Pentinsaari M, Ratnasingham S, Miller SE, Hebert PDN. BOLD and GenBank revisited—do identification errors arise in the lab or in the sequence libraries? PLoS ONE. 2020;15(4):e0231814. https://doi.org/10.1371/journal.pone.0231814.
Polaszek A, Ayshford T, Yahya BE, Fusu L. Wallaceaphytis: an unusual new genus of parasitoid wasp (Hymenoptera: Aphelinidae) from Borneo. J Nat Hist. 2014;48:1111–23. https://doi.org/10.1080/00222933.2013.852264.
Qiu JW, Chan MT, Kwong KL, Sun J. Consumption, survival and growth in the invasive freshwater snail Pomacea canaliculata: does food freshness matter? J Mollus Stud. 2011;77:189–95. https://doi.org/10.1093/mollus/eyr005.
Rama Rao SR, Liew T, Yow YY, Ratnayeke S. Cryptic diversity: two morphologically similar species of invasive apple snail in Peninsular Malaysia. PLoS ONE. 2018;13(5):e0196582. https://doi.org/10.1371/journal.pone.0196582.
Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol Ecol Notes. 2007;7:355–364. https://doi.org/10.1111/j.1471-8286.2006.01678.x.
Rawlings TA, Hayes KA, Cowie RH, Collins TM. The identity, distribution, and impacts of non-native apple snails in the continental United States. BMC Evol Biol. 2007;7:97. https://doi.org/10.1186/1471-2148-7-97.
Senthilkumar K, Rodenburg J, Dieng I, Vandamme E, Sillo FS, Johnson J-M, Rajaona A, Ramarolahy JA, Gasore R, Bayuh BA, Kajiru GJ, Mghase J, Lamo J, Rabeson R, Saito K. Quantifying rice yield gaps and their causes in Eastern and Southern Africa. J Agron Crop Sci. 2020;206:478–90. https://doi.org/10.1111/jac.12417.
Sers CF, Mughal M. Covid-19 outbreak and the need for rice self-sufficiency in West Africa. World Dev. 2020. https://doi.org/10.1016/j.worlddev.2020.105071.
Seuffert ME, Martín PR. Thermal limits for the establishment and growth of populations of the invasive apple snail Pomacea canaliculata. Biol Invasions. 2017;19:1169–1180. https://doi.org/10.1007/s10530-016-1305-0https://ezproxy01.rhul.ac.uk/.
Tamburi NE, Martín PR. Feeding rates and food conversion efficiencies in the apple snail Pomacea canaliculata (Caenogastropoda: Ampullariidae). Malacologia. 2009;51:221–33. https://doi.org/10.4002/040.051.0201.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526
Thiengo SC, Borda CE, Araújo JLB. On Pomacea canaliculata (Lamarck, 1822) (Mollusca; Pilidae: Ampullariidae). Mem I Oswaldo Cruz. 1993;88:67–71.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80.
Wu M, Xie Y. The golden apple snail (Pomacea canaliculata) in China. In: Joshi RC, Sebastian LS, editors. Global advances in ecology and management of golden apple snails. Munoz, Nueva Ecija: PhilRice; 2006. p. 285–98.
Wu J, Meng PJ, Liu MY, Chiu YW, Liu LL. A high incidence of imposex in Pomacea apple snails in Taiwan: a decade after triphenyltin was banned. Zool Stud. 2010;49:85–93.
Yang QQ, Yu XP. A new species of apple snail in the genus Pomacea (Gastropoda: Caenogastropoda: Ampullariidae). Zool Stud. 2019;58:13. https://doi.org/10.6620/ZS.2019.58-13.
Yang QQ, Liu SW, He C, Yu XP. Distribution and the origin of invasive apple snails, Pomacea canaliculata and P. maculata (Gastropoda: Ampullariidae) in China. Sci Rep. 2018;8:1185. https://doi.org/10.1038/s41598-017-19000-7.
Acknowledgements
Grateful thanks are due to Yuen Ting Yeap (CABI) for technical assistance in the molecular identifications. We acknowledge Aidan Emery (Natural History Museum, UK) for helpful suggestions with regard to DNA methodology for snails. Ken Hayes (Bishop Museum, Hawaii, USA) is thanked for guidance on the interpretation of the EF1a sequence. Finally, two anonymous reviewers are thanked for their constructive comments
Funding
This work was undertaken through the Action on Invasives (AoI) programme funded by the United Kingdom (Foreign, Commonwealth & Development Office) and Netherlands (Directorate-General for International Cooperation). CABI is an international intergovernmental organisation, and we gratefully acknowledge the core financial support from our member countries (and lead agencies) including the United Kingdom (Foreign, Commonwealth and Development Office), China (Chinese Ministry of Agriculture), Australia (Australian Centre for International Agricultural Research), Canada (Agriculture and Agri-Food Canada), Netherlands (Directorate-General for International Cooperation), and Switzerland (Swiss Agency for Development and Cooperation). See https://www.cabi.org/about-cabi/who-we-workwith/key-donors/ for full details.
Author information
Authors and Affiliations
Contributions
FM, DC and KV undertook the initial survey and provided detail on the survey methodology; LO did the laboratory analysis of the snail material in the UK and provided the methodological input; GC undertook the molecular processing for identification and supplied the technical information. AB developed the concept, undertook the phylogenetic analyses and wrote the first draft. DD provided the background detail on the invasive species. IR and RD provided funding and supervised the study. AB, DD, IR, AM, SK and RD contributed further edits. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication.
Not applicable.
Competing interests
Several of the authors (i.e. AB, IR, LO, FM, DD, GC, DC and RD) work for CABI, which produces the journal. None of these (or their non-CABI co-authors) had any advance contact with the Editor or with the anonymous Reviewers (and the submission was made using the journal’s double-blind submission process). Aside from this, the authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Buddie, A.G., Rwomushana, I., Offord, L.C. et al. First report of the invasive snail Pomacea canaliculata in Kenya. CABI Agric Biosci 2, 11 (2021). https://doi.org/10.1186/s43170-021-00032-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43170-021-00032-z