Abstract
Background
Wild relatives of wheat are one of the most important genetic resources to use in wheat breeding programs. Therefore, identifying wild relatives of wheat and being aware of their diversity, is undeniably effective in expanding the richness of the gene pool and the genetic base of new cultivars and can be a useful tool for breeders in the future. The present study was performed to evaluate the molecular diversity among 49 accessions of the genera Aegilops and Triticum in the National Plant Gene Bank of Iran using two DNA-based markers, i.e., SSR and ISSR. Also, the present study aimed to examine the relationships among the accessions studied belonging to different genetic backgrounds.
Results
Ten SSR and tan ISSR primers produced 2065 and 1524 polymorphism bands, respectively. The number of Polymorphic Bands (NPB), the Polymorphism Information Content (PIC), Marker Index (MI), and Resolving Power (Rp) in SSR marker was 162 to 317, 0.830 to 0.919, 1.326 to 3.167, and 3.169 to 5.692, respectively, and in the ISSR marker, it was from 103 to 185, 0.377 to 0.441, 0.660 to 1.151, and 3.169 to 5.693, respectively. This indicates the efficiency of both markers in detecting polymorphism among the accessions studied. The ISSR marker had a higher polymorphism rate, MI, and Rp than the SSR marker. Molecular analysis of variance for both DNA-based markers showed that the genetic variation within the species was more than the genetic diversity between them. The high level of genomic diversity discovered in the Aegilops and Triticum species proved to provide an ideal gene pool for discovering genes useful for wheat breeding. The accessions were classified into eight groups based on SSR and ISSR markers using the UPGMA method of cluster analysis. According to the cluster analysis results, despite similarities between the accessions of a given province, in most cases, the geographical pattern was not in accordance with that observed using the molecular clustering. Based on the coordinate analysis, neighboring groups showed the maximum similarities, and distant ones revealed the maximum genetic distance from each other. The genetic structure analysis successfully separated accessions for their ploidy levels.
Conclusions
Both markers provided a comprehensive model of genetic diversity between Iranian accessions of Aegilops and Triticum genera. Primers used in the present study were effective, informative, and genome-specific which could be used in genome explanatory experiments.
Similar content being viewed by others
Background
Wheat is one of the most important staple foods in the world, which is ranked as second among various crops in terms of its area under cultivation and production rate [1]. This crop provides 20% of the total protein and calories needed for human nutrition as well as about 40% of essential micronutrients including zinc, iron, manganese, magnesium, and vitamins B and E for the majority of people whose diet depends on wheat [2]. Global demand for wheat is increasing along with population growth. The statistics show that the demand for wheat and its products will increase up to 50% by 2050 [3]. Producing more food to feed a growing population requires improvement of a more diverse genetic basis of breeding lines and access to new sources of genetic variation for breeding traits, e.g., yield components [4].
On the other hand, one of the most significant factors affecting wheat production is uncertainty in climate patterns [5]. In recent years, climate changes due to abiotic stresses such as drought, heat, and salinity have dramatically impacted the production of crops. Focusing on using new strategies to face the adverse conditions created by these changes and producing resistant/tolerant varieties for cultivation under such climate conditions is one of the basic requirements in breeding programs [6]. Productivity, flexibility, and sustainability of current agricultural systems are essential owing to the land limitation, water, and other inputs of natural resources, competition for arable land, soil degradation, and climate change. And the primary key to sustainable improvement is the use of genetic diversity in plant breeding for sustainable production [7, 8]. Using wild relatives can provide rich and diverse sources of new and ideal alleles for breeders considering the limitation of genetic diversity in modern important crop cultivars for the purpose of adaptation to climate change and other adverse environmental stresses, utilization of available allelic buffer in the gene pool of a given genetic resource germplasm can increase the chance of achieving new allelic diversity in these plant materials [9] Likewise, wheat germplasm is very important to overcome these challenges because native cultivars and wild relatives of wheat have untapped genetic diversity [10,11,12]. Indeed, native cultivars and their wild relatives contain the most appropriate genes due to their genetic capacity and adaptability to physiological conditions and adverse environmental factors, hence providing the genetic diversity required for plant breeders. Therefore, these genetic materials are considered valuable genetic resources, especially in resistance/tolerant to biotic and abiotic stresses, as well as in genetic improvement for other useful traits such as protein quality [13]. Various molecular marker techniques such as AFLP, RAPD, SSR, ISSR, and DArT have recently played a vital role in investigating diversity and evolutionary relationships [14]. The SSR markers are repetitive sequences consisting of one to six nucleotides distributed abundantly and uniformly in the coding and non-coding regions of the eukaryotes genome. These markers are widely used due to their multi-allelic buffering, codominant inheritance, and ease of molecular detection for its variability. The ISSR markers have been shown as efficient markers in genetic diversity studies because of their multilocus, dominance, repeatability, and high polymorphism features [15]. The SSR and ISSR markers have been applied to amplify orthologous sequences in phylogenetically related species in various plants, including wheat [16,17,18], durum wheat [19, 20], barley [21,22,23], triticale [24], sugar beet [25], oat [26], and maize [27].
Two genera Aegilops and Triticum belonging to the Poaceae family are considered the most important genetic resources of wheat. Iran is one of the richest centers of wheat diversity in the Middle East and the Fertile Crescent, so the existence of natural habitats of relative species of Aegilops and Triticum in these regions has led to the creation of one of the wealthiest wheat gene pools in the region [28]. The present study was conducted using SSR and ISSR markers to investigate genetic diversity and to study relationships among various accessions belonging to some species collected from different parts of Iran.
Methods
Plant material and DNA extraction
The plant materials studied in this experiment included 14 accessions of the genus Triticum and 35 accessions of the genus Aegilops from the National Plant Gene Bank of Iran (NPGBI). Control cultivars belonged to the Triticum aestivum which includes five cultivars of Mahoti, Kavir, Rakhshan, Talaei, and Torabi. Plant materials information, along with the genotype code and their geographical characteristics of the place of their collection, are presented in Table 1. The first control, Mahoti as native variety, was selected and introduced from the saline areas around Ardakan in Yazd under saline soil and irrigation conditions. The second control, Kavir, is a saline/drought-tolerant cultivar resulting from the crossing of a native Iranian cultivar resistant to an CIMMYT (International Wheat and Maize Breeding Research Center) line. Three other checks had also a genetic background from CIMMYT. Two checks, i.e., Talaei and Torabi, contain drought-resistance genes from Aegilops tauschii.
The studied accessions were cultivated in Coco peat and perlite substrate and kept in the growth room. Fresh leaf samples of the studied genotypes were collected during stages of 1–2 leaves of growth. Extracting genomic DNA of the desired samples was performed according to the Lodhi et al. [29] method using CTAB (Cethyl Trimethyl Ammonium Bromide) instructions. To ensure the accuracy of the extraction, the quantity and quality of DNA in each sample were measured using a Nanodrop device.
Polymerase chain reaction using markers SSR and ISSR
Ten SSR primers and 10 ISSR primers were used to amplify the genomic DNA of the samples (Table 2). The polymerase chain reaction was prepared in a volume of 10 µl including 3 µl of deionized water, 1 µl of desired primer, 1 µl of Genomic DNA (50 ng concentration), and 5 µl of Master Mix 2X PCR. The polymerase chain reaction thermal cycle includes an initial denaturation step of 5 min at 94°C, followed by 35 cycles of 35 s at 94 °C, binding of 1-min 35 cycles primer at 44–60°C (depending on the type of primer). The duration of primer extension of 1-min 35 cycles, plus the final extension was 10 min. Primer extension of 1-min 35 cycles and the final extension performed at 72 centigrade. We used 1.5% agarose gel with 1X TAE buffer to separate and detect the amplified fragments. This stage is carried out by loading 6 µl of amplified DNA in each well with a voltage of 50 for 50 min. Safe view II was used for staining. Amplified fragments were then checked and detected with a Gel Document device.
Data analysis
The allelic banding patterns was scored as zero and one (as a binary system) for the absence and presence of a band pattern, respectively. Then, various indicators such as polymorphism information content (PIC), resolving power (RP), marker index (MI), total amplified bands (NTB), and number of polymorphic amplified bands (NPB) were used to check and compare the efficiency of various molecular markers and primers. The calculation relationships of each of these indicators are given below:
These relationships, pi, Np, and N, respectively, indicate the allele frequency, the number of polymorphic bands, and the total number of amplified bands from one primer [37].
The use of molecular markers in this research aimed to depict the genetic diversity among the accessions studied in terms of differences attributed to the genus and species, ploidy level, and genomic formula of wheat relatives and the control checks. In order to clarify the relationship between the phenotypic and genotypic data and to select suitable parents for wheat pre-breeding programs, the phenotypic and genotypic diversity of the accessions were compared to each other. Also, the differences within and between the studied populations were performed through molecular variance analysis to explain the differences attributed to the genus and species, ploidy level, and genomic formula of wheat relatives and the check cultivars in this experiment. The genetic structures of 54 genetic samples of Triticum and Aegilops were analyzed using STRUCTURE an Genalex softwares.
Results
Molecular analysis of markers
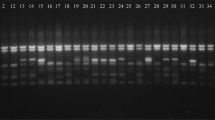
Two examples of the scoring method of the genotypes of wheat relatives of NPGBI after revealing the bands obtained from the markers are shown in Fig. 1. For instance, in the gel “a”, the left and right columns allocated to the 1 kbp Ladder where the position of multi-shape (polymorphic) bands of marker 9 has been demonstrated, and the middle columns were the accessions of NPGBI.
All of the ISSR and SSR primers produced repeatable polymorphic bands. For the ISSR marker, the total produced bands were 1524 bands. The number of polymorphism bands varied from 103 (UBC-840 primer) to 185 (M9 primer), while 152 bands were the average number of observed bands. For each accession, the average number of bands and polymorphism were estimated at 2.345 and 100%, respectively. The average amount of polymorphism information content (PIC), the highest amount of polymorphism, and the lowest amount of polymorphism were 0.41, 0.441 (UBC-857 primer), and 0.377 (M-7 primer), respectively. The highest and lowest index of MI markers were observed in the M-9 (0.660) and UBC- 857 (1.151), respectively. The resolving power of primers (Rp) differed from 3.169 (UBC-840 primer) to 5.692 (M-9 primer). However, its average amount was 4.689.
For SSR markers, the number of total produced bands, average observed bands, and polymorphism were 2065, 206, and 59%, respectively. The number of bands varied from 120 (Xgwm295-7D primer) to 317 (Xgwm192-5D). The average number of bands for each genotype was reported as 2.245 on average. The percentage of polymorphic bands produced by all primers was 100%. The average amount of polymorphism information content (PIC), marker index (MI), and resolving power of primers (Rp) were 0.862, 1.954, and 4.489, respectively. The highest amount was for Xgwm192-5D primer with 317, 3.167, and 6.891 for three parameters of PIC, MI, and Rp. On the other hand, the lowest amount was for Xgwm295-7D primer with 120, 1.064, and 2.609 for three parameters of PIC, MI, and Rp. In general, applied ISSR and SSR primers statistics illustrated the power of the markers in explaining the molecular variations among the genotypes studied.
Genetic diversity among the species
The results of the MANOVA analysis are presented in Table 3. Based on the results, the variance within the populations is very high and includes 74.1, 85.7, and 91.1% of the total variance, respectively. Also, the variance among the populations was significant; it indicated significant genetic differences among the accessions of the groups attributed to species, genomic, and ploidy classification, in terms of polymorphic bands.
The differentiation between populations (Fst-value) average was high for both ISSR and SSR marker systems (Table 4) which a confirmatory for AMOVA results.
The revealed genetic diversity using molecular markers
The statistics obtained from Genalex software were used to explain the situation of the genetic distance of accessions attributed to climatic groups. Estimated genetic alteration parameters, including the average number of observed alleles, the number of effective alleles, Shannon information index, expected heterozygosity, and polymorphism percentage are presented in Table 5. Based on the data of both SSR and ISSR, at the level of species, the highest number of effective allele, Shannon index, and polymorphism percentage were related to aestivum species (0.46, 1.51, and 85%, respectively). Also, at the genome level, it was related to the ABD genome (0.461, 1.508, and 85%, respectively), and at the ploidy level, it was related to the 2n=4X=28 ploidy level (0.479, 1.532, and 93%, respectively). After aestivum, the highest diversity was for the boeoticum and Tauschii species. Moreover, the lowest diversity was observed for the neglecta species, U genome, and 2n=2X=14 ploidy level.
For a better understanding of the similarities and differences in accessions, the Nei similarity coefficient matrix for NPGBI accessions was calculated using ISSR and SSR markers as presented in Table 6. The similarity coefficient among the groups varied from 0.695 (for neglecta and umbellulate species) to 0.956 (for triuncialis and crassa species).
At the levels of genome and ploidy, the Nei similarity coefficient was different from 0.837 (for U and A genomes) to 0.936 (for D and U genomes), and from 0.882 (for 4X and 6X ploidy levels) to 0.975 (for 2x and 4X ploidy level), respectively.
Cluster analysis
The cluster analysis was conducted by the UPGMA method and Jaccard similarity coefficient. According to the data from both SSR and ISSR markers, all accessions were placed in the eight main groups, which in cluster 1, Mahoti and Kavir controls were grouped together. Cluster 2 consisted of 19 accessions belonging to nine species. Cluster 3 included three accessions of cylindrica species. Three controls of aestivum species, namely Rakhshan, Talai, and Tarabi, were included in cluster 4. Five accessions of three species triuncialis, umbellulata, and boeoticum were included in cluster 5. Cluster 6 consisted of nine species belonging to the genus Aegilops, namely six tauschii species, two crassa species and one triuncialis species. Cluster 7 alone contained one boeoticum species. Cluster 8 included 12 genes belonging to the genus Triticum, including eight accessions of boeoticum species, three accessions of urartu species and one accessions of araraticum species (Fig. 2). The species of tauschii with the origin of Gazvin and Kermanshah had the maximum genetic distances (Fig. 3). As demonstrated in the dendrogram of genotypic data, genotypes, like the phenotypic clusters, showed a vast genetic diversity. By carefully studying each cluster’s members, it can be easily observed that in most cases, the molecular pattern was not in accordance with the geographical distribution of accessions, despite the similarities among the accessions of a given province. Some of the genotypes, like quantitative data analysis, were placed at a distance from other members of a similar cluster due to polymorphic band differences. The Garmsar accession, with minimum polymorphic bands, was placed alone in one cluster.
Genetic distance between populations and sub-populations of wheat wild relatives and membership of accessions is presented in Table 7. The maximum and minimum genetic distance observed between A and C, and B and C groups, respectively. The highest number of accessions belonged to the C3 sub-group.
Principal coordinates analysis (PCoA)
To understand the similarities and differences of groups formed based on species and genome formula, coordinate analyses were used, and Figs. 4 and 5 refer to the grouping of the accessions of NPGBI investigated, respectively. In principle coordinates analysis, the two first components explained 76 and 19 percent of the total variation, respectively. As has been found in Fig. 4, the highest genetic similarity is for neighboring groups, while the highest genetic distance is for distant groups.
As shown in Fig. 4, the maximum amount of within species variabilities were observed for the boeticum, aestivum, and triuncialis species, respectively, whereas the minimum levels of within variabilities were observed for the tauschii, cylindrica, and columnaris species, respectively. The neglecta species had the lowest within-group variability; however, its record did not take into account because it only had two samples studied. The most distant accessions belonged to the boeoticum and triuncialis species, whereas the most similar accessions found to be for the species, cylindrica and columnaris. The accessions of urartu with boeoticum as well as those of triuncialis with crassa, columnaris, and cylindrica, and aestivum with boeoticum, triuncialis, and cylindrica demonstrated the highest rate of between-species similarities. Interestingly, tauschii accession had no similarity with other groups except for some crassa accessions.
The A and D genomes had the maximum and the U genome had the minimum variability proportion (Fig. 5). The D genome had the highest similarity with the U genome while did not have any commonality with the A genome. The maximum genetic distance observed between accessions of A and D genomes. The U genome had commonality with all of the genomes studied. The ABD genome (bread wheat) which developed from its wild relatives through evolutionary prosses had a commonality with all others and located in the center of the plot. Additionally, the ABD genome showed high similarity with U and D genomes. The genome D had a wide distribution across both the first and second coordinates.
Structure analysis
The genetic structures of 54 accessions of Triticum and Aegilops were performed and three groups were identified based on maximum-likelihood method (Fig. 6). The ISSR, SSR primers, and both together divided accessions into three groups. These groups correspond to the A, D, and U genomes in wheat wild relatives. Selection for genome A was properly done and all accessions of this genome belonged in a group.
Discussion
In recent years, climate changes have seriously affected agricultural production due to abiotic stresses, such as drought, heat, and salt [38]. These alterations are seriously threatening the productivity of the cultivated wheat [39]. Wheat improvers need to access new genetic resources to support the growing human population demands and to produce more food supplies with higher quality in variable environmental conditions [38]. Wild relatives of wheat are precious resources for useful genetic diversity, which can be used for improving the crops such as wheat [40, 41]. Triticum and Aegilops genomes are the principal parts of the domesticated wheat genetic repository, which contains various species with diverse genomes. The studies revealed that each of the wild relatives has useful alleles, which can be applied in different environmental conditions for cultivated wheat [27]. Therefore, evaluating the genetic diversity within and between the bread wheat wild relatives is necessary before assessing the resistance to biotic and abiotic stresses [33]. Genetic diversity analysis through DNA-based molecular markers has been used as an efficient approach for estimating genome diversity and population structure to utilize in different plants.
In the present research, SSR and ISSR markers were used to investigate within and between species genetic diversity in Aegilops, Triticum, and five bread wheat varieties as control checks. Previously, Nouri et al. [42] used the ISSR marker to evaluate the polymorphism in accessions of Aegilops species. Moreover, Pour-Aboughadareh et al. [43] used SSR marker to investigate the polymorphism in accessions of Aegilops and Triticum species. The efficiency of these markers for designating the polymorphism had been approved. ISSR and SSR primers can target the microsatellites, which can be found plentifully in the plant genome, hence more repeatable markers, as compared to the other markers, e.g., RAPD [44]. Resolution power (Rp) and the polymorphism information content (PIC), in addition to polymorphism percentage, are essential factors of the usefulness of the marker, which are used to compare the efficiency of the marker for genetic analysis. Botstein et al. [45] concluded that primers with 0.25 to 0.50% PIC containbeneficial information for genetic diversity studies. In the present study, the PIC values ranged from 0.377 to 0.919. The average PIC values for ISSR (0.410) and SSR (0.862) indicated the effectiveness of applied primers to demonstrate genetic diversity analysis as well as grouping the accessions of different Aegilops and Triticum species (Fig. 2).
Higher values of MI and Rp for SSR primers validate more clarity and powers of these primers as compared to ISSR primers. Etminan et al. [19] used ISSR and SCoT to detect the genetic diversity in durum wheat genotypes. They concluded that MI and Rp could be the most important indices for determining the effectiveness of a marker. Therefore, ISSR primers proved to be more effective as compared to the SCoT primers. In contrast, Shaban et al. [46] investigated the efficiency of two ISSR and SCoT markers; based on polymorphism percentage, Rp, and MI parameters, SCoT primers were more effective than ISSR primers. Cluster analysis results based on three SCoT, CBDP, and SSR markers, and combined data, showed that the marker of SSR has more efficiency in grouping of Ae. Tauschii, Ae. cylindrica, Ae. crassa, and T. aestivum in two Aegilops and Triticum species [43]. Also, Haque et al. [47] reported 0.69 and 0.73 values for PIC and Nei coefficients for SSR markers in wheat genotypes, respectively. The SSR and ISSR markers had successfully been applied in barley [22] and D genome of Triticum and Aegilops species [48].
In this research, once we applied the information of both markers together, the results of AMOVA showed that the level of the genetic diversity for species (74.1), genome (85.7), and ploidy levels (91.1) of examined accessions were more than the situation that the genetic diversity was estimated for each marker alone. It means both markers’ information could result in a better understanding and a more significant genetic diversity among accessions of the different species, once studying different genomes and ploidy levels. The achieved results correspond to the previous studies, which had reported high levels of diversity in various species of Aegilops and Triticum through different DNA markers [14, 33, 47,48,49]. High values of Fst estimates reflected high proportion of genetic differentiations among the populations investigated. Similar effective differentiation based on SSR data had been reported by Yu et al. [50] in Aegilops tauschii. Also, high values of Fst estimated here also confirmed significant proportion of genetic diversity among populations in the AMOVA analysis.
According to the results based on Shannon information index, number of effective alleles, and polymorphism percentage indices with values of 0.46, 1.51, and 85%, respectively the highest amounts of genetic diversity was found for aestivum species while using two markers at the same time. Likewise, the ABD genome and 2n=4X=28 ploidy level showed the highest genetic diversity bast on above mentioned indices. Moreover, the lowest amount of diversity was for neglecta specie, U genome, and 2n=2X=14 ploidy level. After the aestivum species, species of tauschii and boeoticum had the most genetic diversity. The highest variability observed for ABD genome was due to the richness of allelic variation in the aestivum species whereas the highest variability in tetraploids here was due higher number of 4X accessions as compared to other ploidy level studied in this research. The same reason for the lowest genetic variability showed by neglecta can be addressed to the lowest number of accessions studied in this species. Etminan et al. [19] reported the most observed diversity for aestivum species, followed by Ae. crassa and Ae. cylindrical. The high level of genomic diversity in Aegilops and Triticum species can act as an ideal genetic repository to discover novel genes for modern wheat cultivars [49].
Based on the coordinate analysis, the boeticum, aestivum, and triuncialis species had high within population genetic diversity. Therefore, screening within these populations for abiotic stresses may lead to select promising accession to utilize in wheat improvement programs, while utilizing the species with low genetic diversity could be misleading. According to the coordinate analysis, the primers applied were more effective in separation the boeoticum species than other species. However, Khodaee et al. [33] reported the Iranian accessions of the genetic grouping pattern of Aegilops triuncialis based on the SCoT marker and concluded The differences between markers and primers, primers sequence, or the sample size might cause different results in different studies. Also, principal coordinates analysis revealed that aestivum species (ABD genome) was neighbor and had overlap with other species indicating gene introgression from wild relatives to bread wheat. Therefore, these relatives had high potential to improve new wheat cultivars for adverse environmental conditions.
Structure analysis based on SSR marker data could highly be effective and clearly explain an existing genetic variation among populations [51]. Structure analysis approved PCoA grouping results and A, D, and U genomes separated. This indicates that primers used were genome-specific and could be used in further genome explanatory studies.
Conclusion
In the present study, the molecular results proved the existence of genetic diversity between the studied accessions. Molecular results explained the similarities and differences in the population genetics of the relatives of the National Plant Gene Bank of Iran and led to the identification of polymorphic bands. As a result, these polymorphic bands showed significant correlations with phenotypic grouping. These markers can be utilized to screen wheat genotypes for different traits. Primers used in the present study were effective and genome-specific and could be used in further genome explanatory studies.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- SSR:
-
Simple Sequence Repeats
- ISSR:
-
Inter-simple sequence repeats
- MI:
-
Marker index
- PCoA:
-
Principal coordinate analysis
- RP:
-
Resolving power
References
FAOSTAT.2018. http://faostat3.fao.org/download/Q/QC/E.
Velu G, Singh RP, Huerta J, Guzmán C (2017) Genetic impact of Rht dwarfing genes on grain micronutrients concentration in wheat. Field Crops Res 214:373–377. https://doi.org/10.1016/j.fcr.2017.09.030
Yang X, Tan B, Liu H, Zhu W, Xu L, Wang Y, Fan X, Sha L, Zhang H, Zeng J, Wu D, Jiang Y, Hu X, Chen G, Zhou Y, Kang H (2020) Genetic diversity and population structure of asian and european common wheat accessions based on genotyping-by-sequencing. Front Genet 11:580782
Redden R, Yadav SS, Maxted N, Dulloo ME, Guarino L, Smith P (2015) Crop wild relatives and climate change. Wiley, Hoboken
Mondal S, Sallam A, Sehgal D, Biswal AK, Farhad M, Krishnan NJ, Kumar U, Sukumaran S, Nehela Y (2021) Advances in breeding for abiotic stress tolerance in wheat. In book: Genomic Designing for Abiotic Stress Resistant. Springer, Cereal Crops Publisher
Fita A, Rodrıguez-Burruezo A, Boscaiu M, Prohens J, Vicente O (2015) Breeding and domesticating crops adapted to drought and salinity: a new paradigm for increasing food production. Front Plant Sci 6:978. https://doi.org/10.3389/fpls.2015.00978
Asseng S, Ewert F, Martres P, Rotter RP, Lobell DB, Cammarano D, Kimball BA, Ottman MJ, Wall GW, White JW, Reynolds MP, Alderman PD, Prasad PVV, Aggarwal PK, Anothai J, Basso B, Biernath C, Challinor AJ, de Sanctis G, Doltra J, Fereres E, Garcia-Vila M, Gayler S, Hoogenboom G, Hunt LA, Izaurralde RC, Jabloun M, Jones CD, Kersebaum KC, Koehl A-K, Müller C, Naresh Kumar S, Nendel C, O’Leary G, Olesen JE, Palosuo T, Priesack E, Eyshi Rezaei E, Ruane AC, Semenov MA, Shcherbak I, Stoöckle C, Stratonovitch P, Streck T, Supit I, Tao F, Thorburn PJ, Waha K, Wang E, Wallach DD, Wolf J, Zhao Z, Zhao Y (2015) Rising temperatures reduce global wheat production. Nature Clim Change 5:143–147. https://doi.org/10.1038/NCLIMATE2470
Castañeda-Álvarez NP, Khoury CK, Achicanoy HA, Bernau V, Dempewolf H, Eastwood RJ, Guarino L, Harker RH, Jarvis A, Maxted N, Müller JV, Ramirez-Villegas J, Sosa CC, Struik PC, Vincent H, Toll J (2016) Global conservation priorities for crop wild relatives. Nat Plants 2:1–6. https://doi.org/10.1038/nplants.2016.22
Warschefsky E, Penmetsa RV, Cook DR, Von Wettberg EJ (2014) Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am J Bot 101:1791–1800. https://doi.org/10.3732/ajb.1400116
Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L (2017) Past and future use of wild relatives in crop breeding. Crop Sci 57:1070–1082. https://doi.org/10.2135/cropsci2016.10.0885
Müller T, Schierscher-Viret B, Fossati D, Brabant C, Schori A, Keller B, Krattinger SG (2018) Unlocking the diversity of genebanks: whole-genome marker analysis of Swiss bread wheat and spelt. Theor Appl Genet 131:407–416. https://doi.org/10.1007/s00122-017-3010-5
Milner SG, Jost M, Taketa S, Mazon LR, Himmelbach A, Oppermann M, Weise S, Knüpffer H, Basterrechea M, König P, Schuler D, Sharma R, Pasam RK, Rutten T, Guo G, Xu D, Zhang J, Herren G, Muller T, Krattinger SG, Keller B, Jiang Y, Gonzalez MY, Zhao Y, Habeku A, Färber S, Ordon F, Lange M, Börner A, Graner A, Reif JC, Scholz U, Mascher M, Stein N (2019) Gene bank genomics highlights the diversity of a global barley collection. Nat Genet 51:319–326. https://doi.org/10.1038/s41588-018-0266-x
Pour-Aboughadareh A, Omidi M, Naghavi MR, Etminan A, Mehrabi AA, Poczai P (2020) Wild relative of wheat respond well to water deficit stress: a comparative study of antioxidant enzyme activities and their encoding gene expression. Agriculture 10:425. https://doi.org/10.3390/agriculture10090415
Ghobadi G, Etminan A, Mehrabi AM, Shooshtari L (2021) Molecular diversity analysis in hexaploid wheat (Triticum aestivum L.) and two Aegilops species (Aegilops crassa and Aegilops cylindrica) using CBDP and SCoT markers. J Genet Eng Biotechnol 19:56
Shayan S, Moghaddam Vahed M, Mohammadi SA, Ghassemi-Golezani K, Sadeghpour F, Yousefi A (2020) Genetic diversity and grouping of winter barley genotypes for root characteristics and ISSR markers. Plant Productions 43:323–336
Naghavi MR, Ranjbar M, Zali A, Aghaei MJ, Mardi M, Pirseyedi SM (2009) Genetic diversity of Aegilops crassa and its relationship with Aegilops tauschii and the D genome of wheat. Cereal Res Commun 37:159–167. https://doi.org/10.1556/CRC.37.2009.2.2
Salehi M, Arzani A, Talebi M, Rokhzadi A (2018) Genetic diversity of wheat wild relatives using SSR markers. Genetika 50:131–141. https://doi.org/10.2298/GENSR1801131S
Abbasov M, Brueggeman R, Raupp J, Akparov Z, Aminov N, Bedoshvili D, Gross T, Gross P, Babayeva S, Izzatullayeva V, Mammadova SA, Hajiyev E, Rustamov K, Bikram S-G (2019) Genetic diversity of Aegilops L. species from Azerbaijan and Georgia using SSR markers. Genet Resour Crop Evol 66:453-463. 9
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Noori A, Mahdavian Z (2016) Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip 30:1075–1081. https://doi.org/10.1080/13102818.2016.1228478
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Moradi Z, Mahdavian Z (2017) Evaluation of genetic diversity in a mini core collection of Iranian durum wheat germplasms. J Anim Plant Sci 27:1582–1587
Wang A, Yu Z, Ding Y (2009) Evolution Genetic diversity analysis of wild close relatives of barley from Tibet and the Middle East by ISSR and SSR markers. Biology 332:393–403. https://doi.org/10.1016/j.crvi.2008.11.007
Marzougui S, Kharrat M, Ben Younes M (2020) Assessment of genetic diversity and population structure of tunisian barley accessions (Hordeum vulgare L.) using SSR markers. Acta Agrobot 73:7343
Karamzade F, Arzani A, Mirmohammady-Maibody SAM, Ebrahim F (2021) Genetic diversity of backcross families derived from crossing between cultivated and wild barley using molecular markers. Agric Biotechnol J 13:131–154
Sozen E (2010) Evaluation of ISSR markers to assess genetic variability and relationship among winter triticale (triticosecale wittmack) cultivars. Pak J Bot 42:2755–2763
Devarumath RM, Kalwade SB, Kawar PG, Kawar K, Sushir V (2012) Assessment of genetic diversity in sugarcane germplasm using ISSR and SSR markers. Sugar Tech 14:334–344. https://doi.org/10.1007/s12355-012-0168-7
Targońska M, Bolibok-Brągoszewska H, Rakoczy-Trojanowska M (2016) Assessment of genetic diversity in Secale cereale based on SSR markers. Plant Mol Biol Rep 34:37–51. https://doi.org/10.1007/s11105-015-0896-4
Soliman E, El-Shazly HH, Börner A, Badr A (2021) Genetic diversity of a global collection of maize genetic resources in relation to their subspecies assignments, geographic origin, and drought tolerance. Reprod. Sci 71:313–325. https://doi.org/10.1270/jsbbs.20142
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan AR, Moghaddam M (2018) Assessment of agro-morphological diversity existing in some of Aegilops species. Cereal Res 7:533–549
Lodhi MA, Ye GN, Weeden NF, Reisch BI (1994) A simple and efficient method for DNA extractions from grapevine cultivars and Vitis species. Plant Mol Biol Rep 12:6–13. https://doi.org/10.1007/BF02668658
Röder MS, Korzun V, Gill BS, Ganal MW (1998) The physical mapping of microsatellite markers in wheat. Genome 41(2):278–83
Fathi T, Sohani MM, Samizadeh H, Mehrabi AA (2014) International journal of biosciences. Vol. 4. In: International Network for Natural Sciences (INNS PUB).
Hajiyev ES, Akparov ZI, Aliyev RT, Saidova SV, Izzatullayeva VI, Babayeva SM, Abbasov MA (2015) Genetic polymorphism of durum wheat (Triticum durum Desf.) accessions of Azerbaijan. Russ J Genet 51:863–870
Khodaee L, Azizinezhad R, Etminan A, Khosroshahi M (2021) Assessment of genetic diversity among Iranian Aegilops triuncialis accessions using ISSR, SCoT, and CBDP markers. J Genet Eng Biotechnol 19:5. https://doi.org/10.1186/s43141-020-00107-w
Patil VR, Sapre SS, Singh C, Ahire S, Talati JG (2013) Genetic Diversity of Indian Wheat Cultivars as Revealed by ISSR Markers. Ind J Agric Biochem 26(1):45–50
Thomas KG, Bebeli PJ (2010) Genetic diversity of Greek Aegilops species using different types of nuclear genome markers. Mol Phylogenet Evol 56(3):951–961
Bouziani MC, Bechkri S, Bellil I, Khelifi D (2019) Evaluation of genetic diversity of Algerian Aegilops ventricosa Tausch. using inter-simple sequence repeat (ISSR) markers. World J Environ Biosci 8:1–6
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends in Plant Sci 1(7):215–222. https://doi.org/10.1016/1360-1385(96)86898-1
Pour-Aboughadareh A, Omidi M, Etminan A, Mehrabi AA (2017) The importance of wild wheat germplasm in breeding for resistance to abiotic stresses. Mod Genet 12:489–504
Iizumi T, Ali-Babiker I-EA, Tsubo M, Tahir ISA, Kurosaki Y, Kim W, Gorafi YSA, Idris AAM, Tsujimoto H (2021) Rising temperatures and increasing demand challenge wheat supply in Sudan. Nat Food 2:19–27. https://doi.org/10.1038/s43016-020-00214-4
Kilian B, Dempewolf H, Guarino L, Werner P, Coyne C, Warburton ML (2021) Crop science special issue: adapting agriculture to climate change: a walk on the wild side. Crop Sci 61:32–36. https://doi.org/10.1002/csc2.20418
Sharma S, Schulthess AW, Bassi FM, Badaeva ED, Neumann K, Graner A, Özkan H, Werner P, Knüpffer H, Kilian B (2021) Introducing beneficial alleles from plant genetic resources into the wheat germplasm. Biology 10:982. https://doi.org/10.3390/biology10100982
Nouri A, Golabadi M, Etminan A, Rezaei A, Mehrabi A (2021) Comparative assessment of SCoT and ISSR markers for analysis of genetic diversity and population structure in some Aegilops tauschii Coss. accessions. Plant Genet Resour: Characterization and Utilization 19:375–383. https://doi.org/10.1017/S147926212100040X
Pour-Aboughadareh A, Poczai P, Etminan A, Jadidi O, Kianersi F, Shooshtari L (2022) An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants (Basel, Switzerland) 11:1205. https://doi.org/10.3390/plants11091205
Debnath SC (2009) Development of ISSR markers for genetic diversity studies in Vaccinium angustifolium. Nord J Bot 27:141–148. https://doi.org/10.1111/j.1756-1051.2009.00402.x
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
AS Shaban SA Arab MM Basuoni MS Abozahra Abdelkawy AM. Mohamed MM, (2022) SCoT, ISSR, and SDS-PAGE Investigation of Genetic Diversity in Several Egyptian Wheat Genotypes under Normal and Drought Conditions Int J Agron 54 1 14 https://doi.org/10.1155/2022/7024028
Haque MS, Saha NR, Islam MT (2021) Screening for drought tolerance in wheat genotypes by morphological and SSR markers. J. Crop Sci. Biotechnol 24:27–39. https://doi.org/10.1007/s12892-020-00036-7
Moradkhani H, Mehrabi AA, Etminan A, Pour-Aboughadareh A (2015) Molecular diversity and phylogeny of Triticum-Aegilops species possessing D genome revealed by SSR and ISSR markers. Plant Breed Seed Sci 71:81–95
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2018) Insight into the genetic variability analysis and relationships among some Aegilops and Triticum species, as genome progenitors of bread wheat, using SCoT markers. Plant Biosyst 152:694–703. https://doi.org/10.1080/11263504.2017.1320311
Yu H, Yang J, Cui H, Abbas A, Wei S, Li X (2021) Distribution, genetic diversity and population structure of Aegilops tauschii Coss. in major wheat-growing regions in China. Agriculture. 11(4):311. https://doi.org/10.3390/agriculture11040311
Pandian S, Satish L, Rameshkumar R, Muthuramalingam P, Rency AS, Rathinapriya P, Ramesh M (2018) Analysis of population structure and genetic diversity in an exotic germplasm collection of Eleusine coracana (L.) Gaertn. using genic-SSR markers. Gene 653:80–90
Acknowledgements
Not applicable
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this study. All authors prepared the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jabari, M., Golparvar, A., Sorkhilalehloo, B. et al. Investigation of genetic diversity of Iranian wild relatives of bread wheat using ISSR and SSR markers. J Genet Eng Biotechnol 21, 73 (2023). https://doi.org/10.1186/s43141-023-00526-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-023-00526-5