Abstract
Background
This research provides a comprehensive analysis of QSAR modeling performed on 25 aryl sulfonamide derivatives to predict their effective concentration (EC50) against H5N1 influenza A virus by using some numerical information derived from structural and chemical features (descriptors) of the compounds to generate a statistically significant model. Subsequently, the molecular docking simulations were done so as to determine the binding modes of some potent ligands in the dataset with the M2 proton channel protein of the H5N1 influenza A virus as the target.
Results
In building the QSAR model, the genetic algorithm task was employed in the variable selection of the descriptors which are used to form the multi-linear regression equation. The model with descriptors, RDF100m, nO, and RDF45p, showed satisfactory internal and external validation parameters (R2train = 0.72963, R2adjusted = 0.67169, Q2cv = 0.598, \( {R}_{\mathrm{pred}}^2= \) 0.67295, R2test = 0.6860) which passed the model criteria of acceptability. Docking simulation results of the more potent compounds (ligands 2, 3, and 8) revealed the formation of hydrophobic and hydrogen bonds with the binding pockets of M2 protein of influenza A virus.
Conclusion
The results in this study can help to advance the research in designing (in silico design) and synthesis of more potent aryl sulfonamides derivatives against H5N1 influenza virus.
Similar content being viewed by others
1 Background
Influenza infection is one of the most commonly known acute viral respiratory disease, which is usually spread by the influenza virus. The influenza infection is also called flu, and there are three (3) key types of the influenza virus namely, A, B, and C which are also classified into different subtypes. The circulating seasonal viruses presently are influenza A (H1N1, H3N2) and influenza B viruses [1]. The European Centre for Disease Prevention and Control (ECDPC) in 2019 published a surveillance report on influenza virus characterization which summarizes the percentage of influenza virus detections in the WHO European Region. The report shows that the cumulative detections have increased from 18,049 to 197,027, with type A (99.1%) prevailing over type B (0.9%) viruses, unlike the 2017–2018 season when type B predominated over type A at the beginning of the season [2]. The monthly risk evaluation of influenza at the human-animal boundary published by the World Health Organization in 2019 showed that several influenza A(H5Nx) subtypes continue to be detected in birds in Africa, Europe, and Asia. In addition, the WHO reported that there are over 860 reported cases of humans infected by the H5N1 virus, and 50% among them are dead since 2003, also establishing that H5N1 virus has higher death rate when compared with other influenza A virus subtypes [1]. There are two (2) main classes of anti-influenza which battles with influenza pandemic and epidemic. However, rimantadine and amantadine are M2 proton channel inhibitors that inhibit uncoating of virus-related ribonucleoprotein [3].
The development of more newly potent compounds is very expensive or high-price trial and it is time-consuming. Computational chemistry techniques such as computer-aided drug design (CADD) might save time and reduce the cost of synthesis of the new potent drugs [4]. Ligand-based drug design used quantitative structure-activity relationship (QSAR) approach in designing new compounds from the best statistical model containing some structural features in numerical data (molecular descriptors) which predicts the properties of the compound such as activity, toxicity [5]. The molecular docking simulation is also commonly employed in structure-based drug design (SBDD) which predicts ligand’s conformation and interactions with the active pocket of a protein or enzyme (receptor). This study focused on combining both molecular docking approach and QSAR modeling method to the assessment of 25 aryl sulfonamide derivatives as a novel of H5N1 inhibitors.
2 Methods
2.1 Computer hardware and software
Dell computer system, with processor properties of Intel ® Core i3-6100U CPU Dual @ 2.30 GHz, 12 GB (RAM), was used to carry out this computational study. The software packages installed includes Spartan 14 V 1.1.2 developed by Wavefunction Inc., Material studio software V.8.0, PyRx virtual screening tool, Discovery Studio Visualizer V. 16.1.0, Chemdraw Ultra software V. 12.0.2, PADEL-Descriptor V2.20, Microsoft Excel 2013 version, and some DTC lab softwares.
2.2 QSAR analysis
2.2.1 Collection of dataset and optimization
Twenty-five (25) already synthesized aryl sulfonamides derivatives together with their tested activity concentrations on H5N1 virus were obtained from the literature [3]. The activity concentrations was measured as the concentration that effectively inhibited the virus plaque formation by 50% (EC50) in μM. The H5N1 inhibitory concentration values of these molecules were further converted to the negative logarithmic scale (pEC50 = EC50/106) in order to reduce skewness in the activity values [4]. The chemical structures of the aryl sulfonamide derivatives along with their observed logarithmic effective concentration were shown in Table 1.
The molecular structures of the aryl sulfonamide analogs showed above were properly drawn using ChemDraw Ultra level software V12.0.2, then saved in (*cdx format). Subsequently, the structures were exported to Spartan 14 software so as to compute their equilibrium geometries at ground state with density functional (DFT/B3LYP/6-31G**) in vacuum, starting from the initial molecular geometry [6]. In principle, geometry optimization is an iterative process whereby the energy and its first derivative with respect to all geometrical coordinates are calculated from a guess geometry or starting geometry, then used the information to project new geometry. Thus, the process continues until the lowest energy or optimized structure of the molecule is achieved.
2.2.2 Descriptors computation and data normalization
The twenty-five (25) optimized structures from Spartan 14 were accordingly saved as SDF format, and then exported to PaDEL descriptors software which is a product of Pharmaceutical Data Exploration Laboratory, developed by Yap Chun Wai [6]. This software allows QSAR users to compute diverse molecular descriptors and fingerprints of a molecule, including electrostatic, topological, spatial, autocorrelation, geometrical, constitutional, and thermodynamic descriptors [7]. Data normalization is a technique often applied as part of data preparation and the goal of the normalization is to change the values of numeric columns in the dataset to a common scale, without distorting differences in the ranges of values scaling from 0 to 1 [8].
2.2.3 Data pretreatment and division
PaDEL descriptor output in MS Excel sheet was subjected to the variable reduction method so as to eliminate constant and highly inter-correlated descriptors based on user-specified variance and correlation coefficient cutoff values using Data Pretreatment GUI 1.2, downloaded from Drug Theoretics and Cheminformatics (DTC) Laboratory website. In order to establish a rational selection of training set and test set, the Dataset Division GUI 1.0 software was used by engaging Kennard-Stone’s algorithm division technique [4].
2.2.4 QSAR model generation
In this research, a Small Dataset Modeler version 1.0.0 tool downloaded from the DTC website; https://dtclab.webs.com/software-tools was used to generate a multi-linear regression model (MLR) based on the genetic algorithm technique in variable selection. This tool is very helpful in performing QSAR modeling, especially for small datasets. It employs an exhaustive double cross-validation approach and a set of optimal model selection techniques including consensus predictions for performing the small dataset QSAR modeling [9].
2.2.5 Internal validation
The QSAR model generated was internally validated using cross-validation technique. In essence, this technique provides adequate information about the predictive reliability of the QSAR equation. The leave-one-out cross-validation technique was adopted in this research and the cross-validated Q2cv was evaluated based on expression (4) below [10, 11].
where \( {\overline{Y}}_{tr} \) is the average observed the concentration of training set, Y is the observed concentration, and Ypred is the predicted concentration in the training set respectively.
The squared correlation coefficient (R2) was determined for the comparison between the predicted concentration by the QSAR equation and observed concentrations from the experiment. Furthermore, the R2 values are proportional to the number of descriptors in the model which is not reliable in determining the predictive response of the model. As such, R2 is adjusted which is defined as in;
where p is the number of the descriptor in the equation and n is number of compounds in the training set.
The numerical changes between the R2 and R2adj are less than 0.3 which shows that the number of descriptors used in the QSAR model is acceptable and vice versa [12]. In addition, it is important to note that good R2 values are not enough measures for validating the model. Therefore, more parameters must be established to point out the predictive capability of the models. The cross-validated Q2cv is ordinarily smaller than the R2 value of the QSAR model because of its diagnostic means of evaluating the predictive power of the model [13].
2.2.6 External validation
The optimal combination of the training set and test data (i.e., Compd ID, descriptor matrix, and response) was subjected to MLRplusValidation 1.3 program to internally and externally validate the model generated including the cross-validation method (leave-one-out) and test set validation based on model acceptable criteria. The following statistical features of the test set were proposed by Golbraikh and Tropsha for a robust QSAR model with good predictive potential [7].
- a.
R2pred > 0.6
- b.
\( \frac{r^2-{r}_0^2}{r^2} \) < 0.1
- c.
0.85 < k < 1.15 or 0.85 < k′ < 1.15
where r2 is the squared correlation coefficient between the observed and predicted activities, \( {r}_0^2 \) is the squared correlation coefficient between the predicted and observed activities, and k and k′ are the regression slopes passing through the origin.
where Ypredtest and \( {Y}_{{\mathrm{obs}}_{\mathrm{test}}} \) are the predicted and observed activity of test set compounds respectively. \( {\overline{Y}}_{\mathrm{training}} \) is the average values of observed activity of the training set compounds
2.2.7 Development of applicability domain (AD)
The applicability domain (AD) of the developed model is defined as the chemical space of compound structure and response where the model predictions are highly reliable. This method is used to detect structural and response outliers from the test set and training set respectively [10, 14]. The leverage approach of determining the applicability domain also known as Williams plot was obtained by plotting a scatter plot of standardized residual and leverage values of both training set and test set. By definition, leverage value is determined based on Eq. (4) [15].
where X(i) represents the vector of descriptors of compound (i), X represents the descriptors matrix, and xT represents matrix transpose of X.
The threshold leverage (F*) is given by the equation:
where p is the number of molecules in the training set and m is the number of molecular descriptors used in the model. In addition, compounds with higher leverage scores which are greater than threshold leverage (i > F*) tend to have unreliable predictions. However, compounds whose leverage scores are less than the threshold score (i < F*) and the standardized residuals are not greater than ± 3α (3 standard deviation units) are said to fall within the applicability domain. Similarly, the Euclidean approach of the applicability domain was also determined based on mean distance scores computed by the Euclidean distance. As such, the Uzairu plot was determined by plotting the standardized residuals against normalized mean distance scores whose ranges are from 0 to 1. The normalized mean distance score for training set ranges from 0 which is for least diverse, and 1 which is for the most diverse training set. However, the normalized mean distance score for test compounds with scores outside the range of 0 to 1 is regarded as outliers which imply that the compounds are outside the applicability domain [11].
2.3 In silico docking study
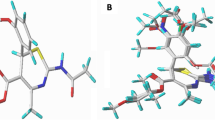
The optimized structures of compounds with the best activity were saved in PDB format (Protein Data Bank), and then docked with the NMR structure of H5N1 virus M2 protein with PDB ID: 2KQT. The docking simulation was carried out using Auto Dock wizard of PyRx virtual screening tool [16, 17]. Subsequently, the docking results were visualized using the Discovery Studio Visualizer v16.1.0.15350 to study the kind of interactions in the stable complex formed. Figure 1 shows 3D structures of the ligand and target respectively.
3 Result
3.1 In silico QSAR analysis
Based on the genetic algorithm of the descriptors, a multi-linear regression model was developed containing three (3) optimum descriptors using Small Dataset Modeler. The selected MLR-GA model is:
Ntrain = 18, SEE = 0.50331, R2train = 0.72963, R2adjusted = 0.67169, PRESS = 3.54655, F = 12.59335, Q2cv = 0.598, average rm2 (LOO) = 0.49284, Δrm2 (LOO) = 0.107, Ntest = 7, \( {R}_{\mathrm{pred}}^2= \) 0.67295, R2test = 0.6860
From the above model (Eq. (6)), it can be deduced that the three (3) most significant descriptors includes - number of oxygen atoms (nO), radial distribution function − 045/weighted by relative polarizabilities (RDF45p), and radial distribution function − 100/weighted by relative mass (RDF100m). Table 2 provides a detailed description of the descriptors in the model. The value of R2train = 0.72963 and R2test = 0.6860 confirms the good extrapolation between the training set and test set respectively. In addition, the QSAR model is robust because of the small difference between R2 and Q2cv (< 0.5%).
The plot of predicted pEC50 against experimental pEC50 values is displayed in Figs. 2, 3, 4, and 5. Visibly, it could be seen that the values of the test sets are in close agreement with the training set values.
3.2 In silico molecular docking
Table 6 summarizes the docking output from PyRx virtual screening and discovery studio visualizer showing the binding affinity scores, distance, and interaction chemistry from the ligand to the receptor or vice versa. Figures 6a–d, 7a–d, 8a–d, and 9a–d showed ligand’s conformation as well as the kind of interaction at molecular level including the hydrogen and hydrophobic interaction types with the active pockets of the M2 proteins of H5N1 influenza virus A in 3D and 2D respectively.
4 Discussion
The key step of building a statistically significant QSAR model is obtaining descriptors that are a portrayal of changes in the structural feature of the compounds. The structural descriptors of all compounds were generated using PaDEL software as stated earlier. A total sum of 1875 descriptors was generated in MS Excel (.csv) format, and the result was exported to a DTC lab software for the normalization and pretreatment process. In the pretreatment process, non-informative and highly inter-correlated descriptors with correlation cutoff greater than 0.7 were removed. The pretreated data were divided into the training set and test set based on Kennard-Stone permutation, where 70% of the dataset (18 compounds) are the training set and the remaining 30% (7 compounds) are the test set. The modeling statistical parameters of the selected model have passed the acceptability criteria proposed by Golbraikh and Tropsha as shown in Table 3. The regression statistics (Table 4) show p value and t values of the model which suggests that the coefficients of the descriptors are statistically significant at 95% confidence interval.
Furthermore, the QSAR model was assessed based on the multi-collinearity among the descriptors by computing the variation inflation factor (VIF) of the three (3) descriptors, which can be computed using the equation:
where R represents the correlation coefficient of the regression between variables in the model. VIF values corresponding to unity depict no inter-correlation among each variable, or if the VIF scores range between 1 and 5, the model is acceptable and stable. But, if the VIF scores are larger than 10, it means that the model in question is unstable and unacceptable [18]. Tables 5 and 6 show the correlation matrix and VIF scores of the descriptors used in forming the selected model. It could be observed that there is no inter-correlation among the descriptors since correlation coefficients are less than 0.5 and VIF scores are not greater than 1 for the three (3) descriptors in the model. The scatter plot of standardized residual versus experimental EC50 (Fig. 3) revealed an unsystematic scattering of data points above and below the baseline of zero data point of standardized residual which signifies the non-existence of systematic error [13]. The scatter plot of standardized residuals against leverage scores also known as Williams plot revealed two (2) response outliers (compounds 2 and 23). This is because there leverage scores are greater than the threshold leverage score of (F*) of 0.66, which may be due to the changes in substitution arrangement of the substituent in the dataset. However, the remaining compounds whose leverage scores are less than the threshold score are said to be within the applicability domain of square area of ± 3. Also, the Uzairu plot showed that all compounds fall within the chemical space of the model which confirmed its predictive capabilities.
In silico docking was conducted on the most active compounds (Comp 2, 3, and 8) with solid-state NMR structure of H5N1 virus M2 protein (target) so as to study the structure-binding relationship of the complex formed. The result showed a binding score of − 5.55 kcal/mol for complex 2, − 5.35 kcal/mol for complex 3, and − 5.22 kcal/mol for complex 8. Ligand 2 formed four (4) hydrogen bond interactions where the thiophene moiety and cyano side chain fill into hydrogen pocket of M2 protein target. The sulfur in thiophene of ligand 2 formed H-bond with HIS37 (2.4406 A°), oxygen of sulfonyl (SO2) serves as H-acceptor in forming H-bond with HIS37 at bond distance of 2.0507 A°, while C-terminal segment of 3081 amino acid residue formed two (2) H-bonds with nitrogen of cyano group and fluoro substituent of benzene ring at distances of 2.2368 A° and 2.4815 A° respectively. Also, the sulfur in sulfonyl group of ligand 2 formed two (2) Π-sulfur interaction with C-terminal segment of HIS37 (4.53254 A°) and D-terminal of HIS37 (5.88837 A°), while sulfur of thiophene moiety also formed Π-sulfur interaction with A-terminal segment of HIS137 at bond distance of 4.42545 A°. Amide-π stacked hydrophobic interaction type was formed between the amide group and delocalized electrons of thiophene ring (ligand 2) at distance 4.2498 A°, also π-orbitals of thiophene fill into hydrophobic pocket of complex 3 with the alkyl group of ILE122 (5.35147 A°). Similarly, ligand 3 formed four (4) hydrogen bond interaction; fluoro substituent of benzene ring served as H-acceptor which interact with A-terminal of GLY34 (2.38373 A°) to form the hydrogen bond. The nitrogen of cyano side chain also formed H-bond with C-terminal segment of 3081 amino acid residue of M2-protein of the H5N1 influenza virus, while sulfur of thiophene moiety and oxygen of the sulfonyl group formed two hydrogen bond interactions with B-terminal of HIS37 and D-terminal of HIS37 at distances of 2.38373 A° and 2.03319 A° respectively. In addition, hydrophobic interaction of amide-π stacked type was formed between the amide group of ILE33 and GLY34 (4.23582 A°) with delocalized Π-electron ring of thiophene. Other interactions in complex 3 includes halogen (fluorine) interaction between fluoro substituent and ALA30 (2.81242 A°); sulfur-X interaction between sulfur of the sulfonyl group and HIS37 (3.2736 A°); three (3) Π-sulfur interaction between sulfur and A: HIS37 (5.86369 A°), D: HIS37 (4.58675 A°), and B: HIS37 (4.34348 A°). Ligand 8 formed only one conventional hydrogen bond with A: HIS37 at bond distance of 2.59672 A°, where oxygen of the sulfonyl group was serving as the H-acceptor. Also, C–H (sigma) interacts with B-terminal segment of HIS37 to form Π-sigma interaction at distance of 3.9007 A°. Π-sulfur interaction was formed between sulfur of thiophene moiety and sulfonyl group with B: HIS37 (4.30161 A°) and C: HIS37 (5.97967 A°) respectively. The presence of hydrophobic and conventional hydrogen bond interaction type in the stable complexes depicts that these ligands (2, 3, and 8) are more potent inhibitors of HN51 influenza virus. Hence, their binding affinity is higher than that of standard drug (amantadine complex) with binding score of − 4.8 kcal/mol, and it formed only four (3) major interaction with the receptor in which three (3) are hydrophobic at different bond distances. The nitrogen of N–H in the amantadine interact with the D-terminal of GLY34 (3.79824 A°) to formed carbon–hydrogen bond interaction, and also the hydrogen formed unfavorable donor–donor interaction as shown in Fig. 9a–d. Furthermore, the molecular docking results in this study are in agreement with Yu et al.’s [3] findings.
5 Conclusion
The present research attempted to use quantitative structure-activity relationship (QSAR) and molecular docking simulations as in silico modeling techniques. The MLR model with molecular descriptors, RDF100m, nO, and RDF45p, obtained from the genetic algorithm task was accepted based on the QSAR model acceptability criteria. The statistical metrics of the descriptors showed that the model is robust and statistically significant. Molecular docking results further revealed the binding modes existing in the stable complex formed between the best ligands with higher activity and the solid-state NMR structure of H5N1 virus M2 protein as the receptor. In addition, the binding affinity scores − 5.55 kcal/mol for complex 2, − 5.35 kcal/mol for complex 3, and − 5.22 kcal/mol for complex 8 are higher than standard drug (amantadine). The outcome of this investigation can help to further the research and development of more potent derivatives of aryl sulfonamides as H5N1 influenza virus inhibitors
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study
Abbreviations
- MLR:
-
Multi-linear regression
- QSAR:
-
Quantitative structure-activity relationship
- NMR:
-
Nuclear magnetic resonance
References
Welkers MRA, Pawestri HA, Fonville JM, Sampurno OD, Pater M, Holwerda M (2019) Genetic diversity and host adaptation of avian H5N1 influenza viruses during human infection. Emerging Microbes and Infections 8(1):262–271
Daniels R, Vicki GJM (2012) Influenza virus characterisation. ECDC - Survillance reports 1:1–7 http://www.ecdc.europa.eu/en/publications/Publications/Influenza-virus-characterisation-September-2012.pdf
Yu Y, Tang Q, Xu Z, Li S, Jin M, Zhao Z (2018) Synthesis and structure-activity relationship study of arylsulfonamides as novel potent H5N1 inhibitors. European Journal of Medicinal Chemistry 159:206–216. https://doi.org/10.1016/j.ejmech.2018.09.065
Abdullahi M, Uzairu A, Shallangwa GA, Mamza P, Arthur DE, Ibrahim MT (2019) In-silico modelling studies on some C14-urea-tetrandrine derivatives as potent anti-cancer agents against prostate (PC3) cell line. Journal of King Saud University - Science. https://doi.org/10.1016/j.jksus.2019.01.008
Ibrahim MT, Uzairu A, Shallangwa GA, Ibrahim A (2018) In-silico studies of some oxadiazoles derivatives as anti-diabetic compounds. Journal of King Saud University – Science. https://doi.org/10.1016/j.jksus.2018.06.006
Adedirin O, Uzairu A, Shallangwa GA, Abechi SE (2018) Optimization of the anticonvulsant activity of 2-acetamido-N-benzyl-2-(5-methylfuran-2-yl) acetamide using QSAR modeling and molecular docking techniques. Beni-Suef University Journal of Basic and Applied Sciences 7(4):430–440. https://doi.org/10.1016/j.jksus.2018.02.009
Sanyal S, Amin SA, Adhikari N, Jha T (2019) QSAR modelling on a series of arylsulfonamide-based hydroxamates as potent MMP-2 inhibitors. SAR and QSAR in Environmental Research 30(4):247–263. https://doi.org/10.1080/1062936X.2019.1588159
Alisi IO, Uzairu A, Abechi SE, Idris SO (2018) Evaluation of the antioxidant properties of curcumin derivatives by genetic function algorithm. Journal of Advanced Research 12:47–54. https://doi.org/10.1016/j.jare.2018.03.003
Ambure P, Gajewicz A, Cordeiro MNDS, Roy K. (2019) Application Note. A new workflow for QSAR model development from small data sets: integration of data curation, exhaustive double cross- validation and a set of optimal model selection techniques, J Chem Inf Model. (just accepted).
Veerasamy R, Rajak H, Jain A, Sivadasan S, Varghese CP, Agrawal RK (2011) Validation of QSAR models - strategies and importance. International Journal of Drug Design and Disocovery. 2(3):511–519, ISSN-0060442980
Arthur DE, Uzairu A, Mamza P, Abechi SE, Shallangwa G (2018) Activity and toxicity modelling of some NCI selected compounds against leukemia P388ADR cell line using genetic algorithm-multiple linear regressions. Journal of King Saud University - Science. https://doi.org/10.1016/j.jksus.2018.05.023
Edache EI, Uzairu A, Abechi SE, Israel E (2015) Investigation of 5,6-dihydro-2-pyrones derivatives as potent anti-HIV agents inhibitors. 5(3):135–49.
Adeniji SE, Uba S, Uzairu A, Arthur DE (2019) A derived QSAR model for predicting some compounds as potent antagonist against Mycobacterium tuberculosis: a theoretical approach. Advances in Preventive Medicine. 2019:1–18. https://doi.org/10.1155/2019/5173786
Tropsha A (2010) Best Practices for QSAR Model Development. Validation, and Exploitation.:476–488
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR and Combinatorial Science 26:694–701. https://doi.org/10.1002/qsar.200610151
Abdulfatai U, Uba S, Umar BA, Ibrahim MT (2019) Molecular design and docking analysis of the inhibitory activities of some α_substituted acetamido-N-benzylacetamide as anticonvulsant agents. SN Applied Sciences 1(5). https://doi.org/10.1007/s42452-019-0512-6
Gholami Rostami E, Fatemi MH (2019) Molecular docking and receptor-based QSAR studies on pyrimidine derivatives as potential phosphodiesterase 10A inhibitors. Structural Chemistry. https://doi.org/10.1007/s11224-019-01353
Driouche Y, Messadi D (2019) Quantitative structure-retention relationship model for predicting retention indices of constituents of essential oils of Thymus vulgaris (Lamiaceae). Journal of the Serbian Chemical Society. 84(4):405–416. https://doi.org/10.2298/JSC180817010d
Acknowledgements
We are thankful to the members of staff in the physical chemistry unit, chemistry department of Ahmadu Bello University, for providing the essential facilities to carry out this research work
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
AU outlined and designed the research work. GAS analyzed and supervised the study. MA handled the computational chemistry software and drafted the manuscript. In addition, all authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Abdullahi, M., Shallangwa, G.A. & Uzairu, A. In silico QSAR and molecular docking simulation of some novel aryl sulfonamide derivatives as inhibitors of H5N1 influenza A virus subtype. Beni-Suef Univ J Basic Appl Sci 9, 2 (2020). https://doi.org/10.1186/s43088-019-0023-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43088-019-0023-y