Abstract
Background
Coronary artery disease (CAD) is the most common kind of heart problem, currently became one of the leading causes of death worldwide and is predicted to persist so for the next 20 years. The global risk factors to CAD include atherosclerosis, genetic predisposition, environment and the lifestyle. This study is aimed to find out the genotypic association of PON1 (rs662) and TNF-α (rs1799964) genes with CAD among North Indian populations. A total of 330 subjects including 175 CAD cases and 155 healthy controls were enrolled in this study. Single nucleotide polymorphisms were analyzed by polymerase chain reaction and restriction fragment length polymorphism (PCR–RFLP) method. χ2 and Student's t-tests were applied for the comparison of alleles and genotype frequencies in cases and controls. Logistic regression analysis was applied to calculate the 95% confidence intervals and odds ratios (OR) for assessing the association of genotype with disease.
Results
The PON1 gene QQ, QR, RR genotypes frequencies were 36.57%, 50.29%, 13.14% in CAD cases and 60%, 38.71%, 1.29% in controls, respectively. OR for the genotype QQ, QR, RR was 0.38, 1.6, 11.57 (P < 0.001, P = 0.035, P < 0.001). The TNF-α gene CC, CT, TT genotypes frequencies in cases were 4.57%, 50.29%, 45.14% and controls 3.23%, 46.45%, 50.32%, respectively. OR for CC, CT, TT genotype was 1.437, 1.166, 0.812 (P = 0.531, P = 0.487, P = 0.347). We found significant difference in the genotype and allele frequencies of PON1 gene between cases and control, while no significant difference was observed in TNF-α gene between cases and control.
Conclusions
The PON1 (rs662) gene polymorphisms were significantly associated with an elevated risk of CAD, while no significant association was observed with TNF-α (rs1799964) gene polymorphism and the risk of CAD.
Similar content being viewed by others
Background
Coronary artery disease (CAD) is the narrowing and blockage of the arteries, usually caused by the buildup of plaque inside the artery walls or atherosclerosis that results to a restriction of blood flow to the heart [1]. According to World Health Organization (WHO) 2015 estimation, 17.7 million people died of CAD that is 31% of all global death [2]. CAD is commonly found in people of every age, but it is more common in progressively older ages [3]. In 2016, the estimated prevalence of CAD in India was estimated to be 54.5 million [4]. One in 4 deaths in India is now because of CAD with ischemic heart disease and stroke responsible for > 80% of this burden. [4]. More than two hundred genetic loci have been identified to be significantly associated with CAD by genome-wide association studies (GWAS) [5].
A calcium-dependent enzyme, human paraoxonase 1 (PON1) is produced by the liver and released into blood [6]. The antioxidant properties possessed by HDL associated PON1 inhibit LDL lipid peroxidation and prevent the atherosclerosis development and CAD [7, 8]. PON1 gene is 26,857-bp long and presents on chromosome 7q21.3 and consists of 8 introns and 9 exons [9]. Among various genetic aberrations, the PON1 (Q192R) gene polymorphism present in the coding region exchanging an arginine (R) for glutamine (Q) at position 192 affect the PON1 activity toward paraoxon. A significant decrease in PON1 activity has been consistently reported in atherosclerosis and related cardiovascular diseases [10, 11]. Recently, several meta-analyses of clinical studies suggested that one of the risk factors for CAD is lower plasma PON1 activity [12, 13].
Tumor necrosis factor (TNF-α), a multi-functional cytokine, is mostly synthesized and secreted by inflammatory cells (monocytes and macrophages) [14]. The human TNF-α gene present on chromosome 6p21.3 within the highly polymorphic major histocompatibility complex (MHC) class III region [15]. Previous study showed that TNF-α is a major participant in the progression of various atherosclerosis complications [16]. TNF-α 308G > A (rs1800629), 857C > T (rs1799724), 238G > A (rs361525), 1031 T > C (rs1799964) and 863C > A (rs1800630) gene polymorphisms are reported so far [17]. Previous studies observed the role of TNF-α rs1799964 gene variant with increased TNF-α secretion and probability of cardiovascular disease [17, 18]. On the basis of these observations, this study was made to examine the role of PON1 and TNF-α genes polymorphism with CAD.
Materials and methods
Subjects
The study was complied with the declaration of institutional ethical/review committee, and written informed consent was obtained from all participants before sample collection. Total 330 subjects including 175 CAD cases (who experienced coronary angiography) and 155 healthy controls were enrolled in the study from January 2017 to December 2019. Blood samples for all the subjects were collected from the Cardiology Unit (Department of Medicine), Era’s Lucknow Medical College & Hospital and from other hospitals of Lucknow. Clinical parameters including age, sex, body mass index (BMI), height, weight, blood pressure, lipid profile, etc., were collected from all the subjects with a standard case report form. Patients with coronary artery diameter’s reduction of > 50% are considered to have CAD. On the basis of number of significant stenotic vessels, all cases were grouped as: angiographically normal vessel (n = 17), 1-vessel (SVD) (n = 42), 2-vessel (DVD) (n = 55) and 3-vessel (TVD) (n = 61). Control subjects include individuals with no previous history of cardiovascular disease or diabetes.
Biochemical estimations
BMI was calculated by Quetelet equation. Random blood sugar (RBS) levels were measured by (glucose oxidase–peroxidase method), serum triglyceride (TG) (glycerol phosphate oxidase–peroxidase amidopyrine method), serum cholesterol (cholesterol oxidase–peroxidase), high-density lipoprotein (HDL) and cholesterol (immunoinhibition) were assessed by XL-300 Transasia Fully Auto Analyzer Transasia, Mannheim, Germany. The measurement of total cholesterol levels and low-density lipoprotein (LDL) was taken by using Friedewald's formula. Standard manufacturer's protocols were followed for all the assays. In this study, each experiment was implemented according to the ethical standards and the declaration of Helsinki.
DNA extraction
Genomic DNA was extracted from blood specimens according to the manufacturers’ protocol using DNA extraction kit (MACHEREY-NAGE, Germany). Extracted DNA was quantified by the Thermo Scientific NanoDrop 2000 Spectrophotometers and stored at − 20 °C until analysis.
Analysis of polymorphisms
PON1 Polymorphism
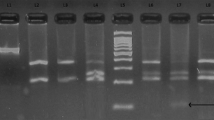
Genotyping of PON1 gene was done by PCR–RFLP method using forward primer 5′ TATTGTTGCTGTGGGACCTGAG 3′ and reverse primer 5′ CACGCTAAACCCAAATACATCTC 3′. Final volume of 20 μL PCR reaction mixture consists of: 10 pmol of each primer, 1 × PCR master mix (Thermo Fisher Scientific Cat no: RR310A), 3 Mm MgCl2, and 2 U Taq polymerase (G Biosciences). PCR conditions were: initial denaturation at 95 °C for 5 min, followed by 35 cycles each consisting of denaturation annealing and extension at 95 °C for 1 min, 61 °C for 1 min and 72 °C for 1 min, respectively, and final extension for 10 min at 72 °C. PCR products were digested with 5 U of restriction enzyme AlwI (New England Biolabs, USA Cat no: R0513S) at 37 °C for 3 h in a total volume of 25 μl, and checked by 3% agarose gel electrophoresis (Fig. 1).
The 3% agarose gel picture of AlwI digested products of PON1 rs664 gene. Lane 1 shows undigested PCR product corresponding to a band of 99 bp, lanes 5 and 8 show QR genotype corresponding to three bands sizes of 99 bp, 66 bp and 33 bp, lanes 3, 6 and 7 show the QQ genotype corresponding to one bands of 99 bp and lane 2 shows RR genotype corresponding to two bands of 66 bp, 33 bp, whereas lane 4 shows 100 bp ladder
TNF-α polymorphism
Genotyping of TNF-α gene was done by PCR–RFLP method using forward primer 5′-TATGTGATGGACTCACCAGGT-3′ and reverse primer 5′-CCTCTACATGGCCCTGTCTT-3′. Final volume of 20 μL PCR reaction mixture consists of: 10 pmol of each primer, 1 × PCR master mix (EmeraldAmp GT PCR master mix), 3 Mm MgCl2, and 2 U Taq polymerase (G Biosciences). PCR conditions were: initial denaturation at 94 °C for 12 min, followed by 35 cycles of annealing and extension at 94 °C for 30 s, 59 °C for 1 min, and 72 °C for 2 min and the final extension at 72 °C for 2 min. PCR products were digested with 3 U of restriction enzyme BbsI (New England Biolabs, USA Cat no: R0539S) at 37 °C for overnight in a total volume of 25 μl and checked by 4% agarose gel electrophoresis (Fig. 2).
The 4% agarose gel picture of BbsI digested products of TNF-α rs1799964gene. Lane 1 shows undigested PCR product corresponding to a band of 264 bp; lanes 2, 4 and 6 show CT genotype corresponding to three bands size of 251 bp, 180 bp and 71 bp; lanes 3 and 7 show the CC genotype corresponding to one bands of 251 bp, whereas lane 8 shows TT genotype corresponding to two band of 180 bp and 71 bp and lane 5 shows 100 bp ladder
Statistical analysis
Values for all the figures were expressed as means ± standard deviation for continuous variables and proportion/percentages for categorical variables. The genotyping data for cases and controls were compared by using Chi-square test and logistic regression analysis. P < 0.05 was represented as statistically significant. All of the statistical tests were performed with the statistical package for the social sciences (SPSS) version 17 software.
Results
Demographic and clinical characteristics
This case–control study comprised 330 subjects including 175 CAD cases and 155 ethnicity matched healthy controls. Clinical and biochemical parameters of CAD cases and controls are shown in Table 1. Demographic, clinical and biochemical characteristics of the studied population were recorded from both cases and controls. The mean ages of CAD cases and control were 54.89 ± 10.47 and 47.30 ± 9.53 years, respectively (P = 0.065). Clinical parameters such as systolic blood pressure (SBP), diastolic blood pressure (DBP), RBS, TG, HDL, LDL (P > 0.05) were significantly higher in cases as compared to controls, whereas no significant difference was observed in BMI (p = 0.797), serum cholesterol (p = 0.215) and VLDL (p = 0.722) in cases compared to the controls (Table 1).
Hardy–Weinberg equilibrium test
PON1 and TNF-α genotype distribution in this study was in line with Hardy–Weinberg equilibrium (all P > 0.05, data not shown).
Genetic polymorphism Analysis
PON1 polymorphism analysis (rs662)
The PON1 gene QQ, QR, RR genotypes frequencies were 36.57%, 50.29%, 13.14% in CAD cases and 60%, 38.71%, 1.29% in healthy controls, respectively. Odds ratio (OR) for QQ, QR, RR genotype was 0.38, 1.6, 11.57 (P < 0.001, P = 0.035, P < 0.001). The frequencies of Q and R alleles were 61.71% and 38.29% in cases as compared to 79.35% and 20.65% in the controls. OR for Q was 0.419 (P < 0.001), and for R allele OR = 2.384 (P < 0.001) (Table 2). The frequency of QQ and RR genotype of PON1 gene was found to be highly significant in cases with 0.3 and 11 fold higher risk of CAD (P < 0.0001) in comparison with control. Similarly, the frequency of Q and R allele of PON1 gene was also found to be highly significant in cases with 0.4 and twofold higher risk of CAD (p ≤ 0.0001).
TNF-α polymorphism analysis (rs1799964)
The TNF-α gene CC, CT, TT genotypes frequencies were 4.57%, 50.29%, 45.14% in CAD cases and 3.23%, 46.45%, 50.32% in healthy controls, respectively. OR for CC, CT, TT genotype was 1.437, 1.166, 0.812 (P = 0.531, 0.487, 0.347). The frequencies of C and T alleles were 29.71% and 70.29% in CAD cases as compared to 26.45% and 73.55% in the controls. OR for C was 1.175 (P = 0.352), and for T allele OR = 0.851 (P = 0.352) (Table 2).
Discussion
CAD has become the major cause of death worldwide, accounting for approximately 30% of deaths and the most common health problem in India [19]. As with most complex diseases, the risk of CAD development in an individual is affected by the genetic and lifestyle factors [20]. The associations of 163 genetic loci with CAD have been revealed so far by GWAS [21]. Our previous studies have also shown the association of gene polymorphism with CAD [22, 23]. Due to the presence in noncoding regions, most of the gene variants are associated with CAD resulting into the development of disease by regulating gene expression [24].
PON1 is a calcium-dependent antioxidant glycoprotein and is regarded as a cardiovascular protective factor. Clinical studies suggested that the lower plasma PON1 activity is significantly associated with increased CAD risk [13, 25]. Approximately 160 polymorphisms of the PON1 gene have been discovered so far, either present in exonic (coding sequences) or intronic (noncoding sequences) region and the regulatory parts of genes [26]. Majority of them include single-nucleotide polymorphisms; however, those in the coding regions were extensively studied, i.e., Q192R (rs662) and L55M (rs854560) [27, 28]. The antioxidative potential of Arg192 may disturb by the exchange of glutamine (Q) to arginine (R)Q in 192R polymorphism and result into the development of CAD [29]. There are different reports suggesting the distribution of Q and R allele in different ethnic populations. Among cases, we found Q and R to be major and minor alleles at codon 192 in the PON1 gene, respectively. The distribution of Q and R allele of the PON1 gene in the present study tended to be closer to that observed in Egyptian [32, 34], Chinese [35] and Asian Indians [36], but differed from South Indian [33] populations where the R allele was predominant (Table 3). The present study showed the significant increase in the QR and RR genotypes frequency of PON1 Q192R polymorphism in cases compared to the controls, hence, indicating that it plays a crucial role in the pathogenesis of CAD. Studies examined the role of the Q192R polymorphism with CAD risk observed inconsistent results. Several studies have shown the positive correlation of Q192R polymorphism with CAD risk [31,32,33,34,35,36], whereas others have not [30]. The PON1 gene Q192R polymorphism contains the ability to inhibit LDL oxidation; hence compared with 192Q allele, the 192R allele is less effective for the prevention LDL oxidation [37]. Thus, R allele’s carriers are more susceptible to develop cardiovascular diseases than Q allele’s carriers. In the present study, significant association was observed between Q and R allele of PON1 gene with CAD (P < 0.001), where Q allele confers the increase risk while R allele with decrease risk of CAD. We have observed that the PON1 QR genotype frequency was 50.29% in CAD cases which is similar to Asian Indian 44.9%, South Indian 51.5% CAD cases [33, 36]. PON1 QQ genotype frequency was 36.57% in our study which is significantly higher compare to South Indian 28% [33]. Further studies including larger sample size from different ethnic groups required to examine the association of PON1 gene polymorphisms in the development of CAD.
One of the most typical pro-inflammatory cytokines TNF-α was suggested by many studies as a major contributor in the atherosclerosis development, complications and progression [38]. Relatively limited studies have been done so far based on the relation of TNF-α-1031T > C (rs1799964) gene polymorphisms and CAD. Hence, this study was carried out to examine the role of TNF-α gene polymorphisms with CAD among North Indian population. Our findings suggest no significant association of TNF-α gene-1031T > C polymorphism with CAD risk in the study population. The TNF-α-1031C allele frequency was 26% in control versus 30% in cases (P = 0.46), indicating that the − 1031C allele is not a risk factor of CAD in our population. This is in accordance with the results of Asifa et al. and Ghazouani et al., demonstrating no significant association between the TNF-α gene polymorphism at − 1031 T > C promoter sequence and CAD among Pakistani and Chinese, respectively [39, 41]. The present study observed the distribution of − 1031 T and − 1031C allele of the TNF-α gene relatively similar to Pakistani population [39] but slightly differed from that of African [40] and Chinese [41] [Table 3]. The frequency of TNF-α TT genotype was 45% in CAD cases which is lower compared to Pakistani population 57% [39].
To the best of our knowledge, this is the first study to look at the relationship between PON1 (rs662) and TNF-α (rs1799964) genes polymorphism in patients with CAD. Further study on a larger cohort of cases with CAD is needed to validate this study.
Conclusions
Our results suggest that the PON1 (rs662) gene polymorphisms were associated with an increased risk of CAD, whereas TNF-α (rs1799964) gene polymorphism was not conferring any risk of CAD. Thus, PON1 (rs662) gene polymorphism may be a useful tool for diagnosis, prognosis, and prediction of the disease and may have an influence on more effective and specific treatment against the CAD.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BMI:
-
Body mass index
- CAD:
-
Coronary artery disease
- CI:
-
Confidence interval
- DBP:
-
Diastolic blood pressure
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- HDL:
-
High-density lipoprotein
- LDL:
-
Low-density lipoprotein
- MHC:
-
Major histocompatibility complex
- OR:
-
Odd ratio
- P:
-
P-value
- PCR:
-
Polymerase chain reaction
- PON1 :
-
A calcium-dependent enzyme, human paraoxonase 1
- RFLP:
-
Restriction fragment length polymorphism
- SBP:
-
Systolic blood pressure
- SNP:
-
Single nucleotide triphosphate
- SPSS:
-
Statistical package for the social sciences
- TNF-α :
-
Tumor necrosis factor
- WHO:
-
World Health Organization
- χ2:
-
Chi-square test
References
Sanchis-Gomar F, Perez-Quilis C, Leischik R et al (2016) Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med 4:256
WHO. http://www.who.int/mediacentre/factsheets/fs241/en/ (2014).
Curtis AB, Karki R, Hattoum A, Sharma UC (2018) Arrhythmias in patients ≥ 80 years of age: pathophysiology, management, and outcomes. J Am Coll Cardiol 71:2041–2057
India State-Level Disease Burden Initiative CVD Collaborators. The changing patterns of cardiovascular diseases and their risk factors in the states of India: the Global Burden of Disease Study 1990–2016.Lancet Glob Health. 2018; 6:e1339–e1351.
Nikpay M et al (2015) comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47:1121–1130
Mackness M, Mackness B (2015) Human paraoxonase-1 (PON1): Gene structure and expression, promiscuous activities and multiple physiological roles. Gene 67:12–21
Litvinov D, Mahini H, Garelnabi M (2012) Antioxidant and anti-inflammatory role of paraoxonase 1: implication in arteriosclerosis diseases. N Am J Med Sci 4:523–532
Soran H, Schofield JD, Durrington PN (2015) Antioxidant properties of HDL. Front Pharmacol 6:764–772
Primo-Parmo SL, Sorenson RC, Teiber J et al (1996) The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics 33:498–507
Macharia M, Hassan MS, Blackhurs D et al (2012) The growing importance of PON1 in cardiovascular health: a review. J Cardiovasc Med (Hagerstown) 12:443–453
Taskıran P, Cam SF, Sekuri C et al (2009) The relationship between paraoxanase gene Leu-Met (55) and Gln-Arg (192) polymorphisms and coronary artery disease. Turk Kardiyol Dern Ars 37:473–478
Wang M, Lang X, Cui S et al (2012) Quantitative assessment of the influence of paraoxonase 1 activity and coronary heart disease risk. DNA Cell Biol 31:975–982
Zhao Y, Ma Y, Fang Y et al (2012) Association between PON1 activity and coronary heart disease risk: a meta-analysis based on 43 studies. Mol Genet Metab 105(1):141–148
Mellick GD (2007) TNF Polymorphism and Cardiovascular Disease: TNF gene polymorphism and quantitative traits related to cardiovascular disease: getting to the heart of the matter. Eur J Hum Genet 6:609–611
Hajeer AH, Hutchinson IV (2001) Influence of TNF alpha gene polymorphisms on TNF alpha production and disease. Hum Immunol 62:1191–1199
Goodwin BL, Pendleton LC, Levy MM et al (2007) Tumor necrosis factor-α reduces argininosuccinate synthase expression and nitric oxide production in aortic endothelial cells. Am J Physiol Heart Circ Physiol 293(2):1115–1121
Hernández-Díaz Y, Tovilla-Zárate CA, Juárez-Rojop I et al (2016) Association between CRP and TNF-α genes variants and cardiovascular heart disease in a mexican population: protocol for a case-control study. Int J Environ Res Public Health 13(1):103
Cui G, Wang H, Li R, Zhang L et al (2012) Polymorphism of tumor necrosis factor alpha (TNF-alpha) gene promoter, circulating TNF-alpha level, and cardiovascular risk factor for ischemic stroke. J Neuroinflammation 9:1742–2094
Singh RB, Sharma JP, Rastogi V et al (1997) Prevalence of coronary artery disease and coronary risk factors in rural and urban populations of north India. Eur Heart J 18(11):1728–1735
Khera AV, et al. Genetic risk, adherence to a healthy lifestyle, and risk of coronary disease. NEJM. 2016.
Erdmann J, Kessler T, Munoz Venegas L et al (2018) A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res 385:526
Raza ST, Singh SP, Rizvi S, Zaidi A, Srivastava S, Hussain A et al (2021) Association of eNOS (G894T, rs1799983) and KCNJ11 (E23K, rs5219) gene polymorphism with coronary artery disease in North Indian population. Afr Health Sci 21(3):1163–1171
Raza ST, Abbas S, Eba A et al (2018) Association of COL4A1 (rs605143, rs565470) and CD14 (rs2569190) genes polymorphism with coronary artery disease. Mol Cell Biochem 445(9859):1–6
Brænne I et al (2015) Prediction of causal candidate genes in coronary artery disease loci. Arterioscler Tromb Vasc Biol 35:2207–2217
Kowalska K, Socha E, Milnerowicz H (2015) The role of paraoxonase in cardiovascular diseases. Ann Clin Lab Sci 45(2):226–233
Costa LG, Vitalone A, Cole TB et al (2005) Modulation of paraoxonase (PON1) activity. Biochem Pharmacol 69:541–550
Draganov DI, La Du BN (2004) Pharmacogenetics of paraoxonases: a brief review. Naunyn Schmiedebergs Arch Pharmacol 369:78–88
Costa LG, Cole TB, Jarvik GP et al (2003) Functional genomic of the paraoxonase (PON1) polymorphisms: effects on pesticide sensitivity, cardiovascular disease, and drug metabolism. Annu Rev Med 54(1):371–392
Serrato M, Marian AJ (1995) A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest 96(6):3005–3008
Bayrak A, Tülin Bayrak A, Lale Tokgözoglu S et al (2012) Serum PON-1 activity but not Q192R polymorphism is related to the extent of atherosclerosis. J Atherosclerosis Thromb 19(4):376–384
Chen H, Ding S, Zhou M et al (2018) PON1 L55M and Q192R gene polymorphisms and CAD risks in patients with hyperlipidemia: clinical study of possible associations. Herz 43(7):642–648
Mohamed RH, Mohamed RH, Karam RA et al (2010) The relationship between paraoxonase1-192 polymorphism and activity with coronary artery disease. ClinBiochem 43:553–558
Matam K, Khan I, Hasan Q, Rao P (2014) Coronary artery disease and the frequencies of MTHFR and PON1 gene polymorphism studies in a varied population of Hyderabad, Telangana region in south India. J King Saud Univ Sci 52:143–150
El-Lebedy D, Kafoury M, Abd-ElHaleem D et al (2014) Paraoxonase-1 gene Q192R and L55M polymorphisms and risk of cardiovascular disease in Egyptian patients with type 2 diabetes mellitus. J Diabetes Metab Disord 13:125
Chen H, Shahein MM, Mochiah MB et al (2017) PON1 L55M and Q192R gene polymorphisms and CAD risks in patients with hyperlipidemia. Herz 43:642–648
Sumanpreet K, Gurjit K, Bhattib R et al (2018) Paraoxonase 1 Gene Polymorphisms (Q192R and L55M) are associated with coronary artery disease susceptibility in Asian Indians. Int J Diabetes Metab 24:38–47
Regieli JJ, Jukema JW, Doevendans PA et al (2009) Paraoxonase variants relate to 10-year risk in coronary artery disease: impact of a high-density lipoprotein-bound antioxidant in secondary prevention. J Am Coll Cardio 54:1238–1245
Goodwin BL, Pendleton LC, Levy MM et al (2007) Tumor necrosis factor-α reduces argininosuccinate synthase expression and nitric oxide production in aortic endothelial cells. Am J Physiol Heart Circulatory Physiol 293(2):1115–1121
Asifa GZ, Liaquat A, Murtaza I et al (2013) Tumor necrosis factor-alpha gene promoter region polymorphism and the risk of coronary heart disease. Sci World J 5:1–5
Ghazouani L, Khalifa SB, Abboud N et al (2009) 308G>A and -1031T>C tumor necrosis factor gene polymorphisms in Tunisian patients with coronary artery disease. Clin Chem Lab Med 47:1247–1251
Cui G, Wang H, Li R et al (2012) Polymorphism of tumor necrosis factor alpha (TNF-alpha) gene promoter, circulating TNF-alpha level, and cardiovascular risk factor for ischemic stroke. J Neuroinflammation 9:1742–2094
Acknowledgements
We are thankful for the study supported as an intramural grant from the Era’s Lucknow Medical College and Hospital, Lucknow, Uttar Pradesh, India.
Funding
The study was supported by the intramural grant from the Era’s Lucknow Medical College and Hospital, Lucknow, Uttar Pradesh, India.
Author information
Authors and Affiliations
Contributions
ST and SA have done overall search and compilation of data. IAW helps in the collection of sample. AE helped in the analysis of data. FM has done overall supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Protocol and procedures in the study complied with the declaration of Institutional Ethical/ Review Committee (Ref no. ELMC &H/R-Cell-/2019/24) of Era’s Lucknow Medical College and Hospital, Lucknow.
Consent for publication
Written informed consent for publication was obtained from all the participants before sample collection.
Competing interests
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Raza, S.T., Abbas, S., Wani, I.A. et al. Clinical implications of PON1 (rs662) and TNF-α (rs1799964) genes polymorphism in patients with coronary artery disease. Egypt J Med Hum Genet 23, 107 (2022). https://doi.org/10.1186/s43042-022-00318-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-022-00318-5