Abstract
Background
Colorectal cancer (CRC) is the third most common cause of cancer-related deaths which contributes to a significant public health problem worldwide with 1.8 million new cases and almost 861,000 deaths in 2018 according to the World Health Organization. It exhibits 7.4% of all diagnosed cancer cases in the region of the Middle East and North Africa. Molecular changes that happen in CRCs are chromosomal instability, microsatellite instability (MSI), and CpG island methylator phenotype. The human RAS family (KRAS, NRAS, and HRAS) is the most frequently mutated oncogenes in human cancer appearing in 45% of colon cancers. Determining MSI status across CRCs offers the opportunity to identify patients who are likely to respond to targeted therapies such as anti-PD-1. Therefore, a method to efficiently determine MSI status for every cancer patient is needed.
Results
KRAS mutations were detected in 31.6% of CRC patients, namely in older patients (p = 0.003). Codons 12 and 13 constituted 5/6 (83.3%) and 1/6 (16.7%) of all KRAS mutations, respectively. We found three mutations G12D, G12C, and G13D which occur as a result of substitution at c.35G>A, c.34G>T, and c.38G>A and have been detected in 4/6 (66.6%), 1/6 (16.7%), and 1/6 (16.7%) patients, respectively. Eleven (57.9%) patients had microsatellite instability-high (MSI-H) CRC. A higher percentage of MSI-H CRC was detected in female patients (p = 0.048). Eight patients had both MSI-H CRC and wild KRAS mutation with no statistical significance was found between MSI status and KRAS mutation in these studied patients.
Conclusion
In conclusion, considering that KRAS mutations confer resistance to EGFR inhibitors, patients who have CRC with KRAS mutation could receive more tailored management by defining MSI status. MSI-high patients have enhanced responsiveness to anti-PD-1 therapies. Thus, the question arises as to whether it is worth investigating this association in the routine clinical setting or not. Further studies with a larger number of patients are needed to assess the impact of MSI status on Egyptian CRC care.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is the third most common cause of cancer-related deaths which contributes to a significant public health problem worldwide with 1.8 million new cases and almost 861,000 deaths in 2018 according to the World Health Organization. It exhibits 7.4% of all diagnosed cancer cases in the region of the Middle East and North Africa [1]. Globally, the highest CRC incidence rates are in Australia, New Zealand, Europe, and North America, and the lowest rates are detected in Africa and south Central Asia [2]. Despite the fact that the overall incidence of CRC in teenagers and young adults is low, it has seen an expansion over the past decades. This is caused by a lack of routine screening in the young population and lifestyle issues such as obesity and dietary factors [3]. Molecular changes that happen in CRC might be classified into three principals groups: (1) chromosomal instability (CIN), (2) microsatellite instability (MSI), and (3) CpG island methylator phenotype (CIMP) [4]. The human RAS family (KRAS, NRAS, and HRAS) is the most frequently mutated oncogenes in human cancer appearing in 90% of pancreatic, 35% of lung, and in 45% of colon cancers. Mutation in Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) is the most important widely used molecular test in the CRC metastatic settings due to the presence of this mutation which can predict a lack of benefit from epithelial growth factor receptor (EGFR)-targeted antibodies. These KRAS mutations are highly frequent in the codons 12, 13, 59, 61, and 146 [1, 5]. B-Raf proto-oncogene, serine/threonine kinase (BRAF) gene mutations are detected in 15% of all cancers, and the V600E mutation is detected in 10% of all CRCs. However, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) gene is implicated in the PI3K/Akt pathway and is mutated in ~ 10–20% of CRCs where ~ 80% mutations are found in two hotspot areas in exons 9 and 20. Also, the patients with mutations in BRAF and PIK3CA have shown a negative response to EGFR inhibitor therapy [1]. Phosphatase and tensin homolog (PTEN) is located in 10q23.31, and its main function is to block the PI3K pathway by dephosphorylating phosphatidylinositol (PI) 3,4,5-triphosphate into PI-4,5-bisphosphate thus opposing PI3K function. PTEN is found mutated in 3.86% of CRC patients. Finally, tumor protein p53 (TP53) is located in 17p13.1. Somatic mutations in the TP53 gene are the most common genetic changes found in human cancer occurring in about 50% of CRCs with the most frequently observed mutations between exons 5 and 8 at codons 175, 213, 245, 248, 273, and 282 [1, 4]. Microsatellites, also known as short tandem repeats (STRs), are small (1–6 bp) repeating stretches of DNA scattered all through the whole genome (both in coding and non-coding regions) and account roughly for 3% of the human genome. Because of their repeated structure, microsatellites are subjected to a high mutation rate. Microsatellite instability (MSI) in tumor DNA is defined as the presence of alternate sized repetitive DNA sequences that are absent in the corresponding germline DNA. MSI is a molecular phenotype due to a defective DNA mismatch repair system [6]. DNA mismatch repair (MMR) system corrects erroneous insertion, deletion, and base-base mismatches generated during DNA replication and recombination that have escaped the proofreading process [6]. Tumors are classified as MMR-deficient (dMMR) if they have somatic or germline mutations. There are several methods to detect MSI, the Bethesda panel of microsatellite markers comprising 2 mononucleotide repeat markers (BAT25 and BAT26) and three dinucleotide repeat markers (D2S123, D5S346, and D17S250). If at least two or more microsatellite markers are mutated, the tumor is considered MSI-high (MSI-H); if just one is mutated, the tumor is defined as MSI-low (MSI-L); and if none of the examined loci exhibits instability, the tumor will be considered microsatellite stable (MSS). A few years later, it was discovered that mononucleotide markers have a better sensitivity and specificity as the dinucleotide markers have a polymorphic nature. So, the uses of panels containing more mononucleotide markers have been expanded due to their higher sensitivity and specificity in the diagnosis of MSI in CRCs [7]. MSI has been associated with improved prognosis as MSI-high cancers are a genetically defined subset of cancers with the potential for enhanced responsiveness to anti-PD-1 therapies. Determining MSI status across cancer types offers the opportunity to identify patients who are likely to respond while avoiding unnecessary toxicities for patients identified as unlikely to respond [8]. Therefore, a method to efficiently determine MSI status for every cancer patient is needed.
Study objectives
Our objective was to demonstrate the clinicopathologic features of Egyptian CRC patients together with identification and correlation with somatic genetic changes and the MSI status in those patients.
Methods
Study population and sample collection
The present study included 19 newly diagnosed fresh surgical tumor specimens from 19 CRC patients attending to the outpatient clinic of our department. All patients were subjected to surgical resection where the surgeon removed the part of the colon that contains the tumor mass, along with a margin of normal tissue on either side of the mass. After gross examination, macro dissection was done. Then, hematoxylin and eosin (H&E) staining was performed on serial sections. The tissue area representing the “tumor” which contains the highest numbers of cancer cells was identified and “normal” which contains no malignant tissue identified and separated. Healthy control tissue was included in our study for the interpretation of MSI results.
Patients were selected according to the following criteria:
Inclusion criteria:
-
Age group: adulthood CRC patients
-
Gender: males and females
-
Pathologically proved colorectal adenocarcinoma.
-
Stages II, III, and IV
-
Newly diagnosed cases attending the outpatient clinic
Exclusion criteria:
-
Patients with double malignancy
-
Known HIV-positive or AIDS-related illness
-
Women who are pregnant or breastfeeding
Our study was exploratory in nature and performed in the molecular lab of our institution, in the period from March 2018 to February 2019. The study was approved by our department Institutional Review Board (IRB)-3-2018. All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments (GCP guidelines) or comparable ethical standards.
DNA extraction
Genomic DNA was extracted from tissues with the QIAamp DNA Mini kit (Qiagen, USA) as reported by the manufacturer and was eluted in 60 μl volume. The extracted DNA samples were measured utilizing the Qubit dsDNA High Sensitivity (HS) assay kit (Life Technologies, Fisher Scientific, USA). A quantitative PCR (qPCR) reaction was performed to conclude the amplifiability of the extracted gDNA samples and to predict assay success. All samples with ΔCq value below or equal to 2 can be selected for further use.
Library preparation
Libraries were prepared using TruSight Tumor 15 kit according to the manufacturer’s protocol (Illumina, Inc., San Diego, CA). TruSight Tumor 15 uses next-generation sequencing (NGS) to assess 15 of the most commonly mutated significant genes in CRC in a single assay: AKT serine/threonine kinase 1 (AKT1), G protein subunit alpha 11 (GNA11), NRAS, BRAF, G protein subunit alpha q (GNAQ), platelet-derived growth factor receptor alpha (PDGFRA), EGFR, KIT proto-oncogene receptor tyrosine kinase (KIT), PIK3CA, erb-b2 receptor tyrosine kinase 2 (ERBB2), KRAS, ret. proto-oncogene (RET), forkhead box L2 (FOXL2), MET proto-oncogene, receptor tyrosine kinase (MET), and TP53. Library quality was checked out by 2100 Bioanalyzer utilizing the Bioanalyzer DNA 1000 reagents and chips (Agilent Technologies, Santa Clara, CA). Successful library amplification was estimated when the expected PCR product size is ~ 350 bp. Before sequencing, the libraries together with PhiX control library were normalized following the manufacturer’s protocol, and equal volumes were pooled to constitute the terminal sequencing library. The TruSight Tumor 15 sequencing panel achieves limits of detection of 5% variant allele frequency across 250 amplicons with 93.5% of bases covered at ≥ 500× [9].
Sequencing and data analysis
Sequencing was accomplished utilizing MiSeqDx device (Illumina) with a 2 × 151 bp read length and a total time of 27 h which involve cluster generation, sequencing, and base calling on the MiSeqDx system. Specifications based on Illumina PhiX control library support cluster densities between 1200 and 1400 k/mm2 clusters passing filter for v3 chemistry. The quality scores > 80% bases higher than Q30 at 2 × 151 bp. Image processing and VCF file generation were further analyzed; we then annotated the variants using VariantStudio™ software version 3. This software determined the numerical identifier for each variant in the COSMIC database, if the genomic variant position overlapped a variant recorded in COSMIC. The COSMIC ID linked to the COSMIC page associated with the identifier to determine information on whether that variant has been detected before in any cancer patient or a novel one. Synonymous variants and non-coding regions were filtered out. Mutations with low depth, which indicate ≤ 50× depths, were filtered out. And mutations with ≤5% variant allele frequency were filtered out. Variant quality which is one parameter of the variant call format (VCF) < 80% was filtered out.
MSI analysis
We evaluated the MSI status using three mononucleotide repeat markers (BAT25, BAT26, and NR27). The kit was purchased from Qiagen Co., with primer sequences as follows:
BAT-25 (product size, 114 bp): 5′-CTCGCCTCCAAGAATGTAAGT-3′; 5′-CTATGGCTCTAAAATGCTCTGTTC-3′
BAT-26 (product size, 122 bp): 5′-TGACTACTTTTGACTTCAGCC-3′; 5′-AACCATTCAACATTTTTAACCC-3′
NR-27 (product size, 89 bp): 5′-AACCATGCTTGCAAACCACT-3′; 5′-CGATAATACTAGCAATGACC-3′.
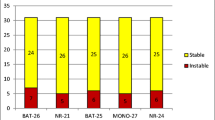
Amplification reactions (20 μl) were prepared with 10–100 ng extracted DNA, 1× PCR buffer, 1.5 mM MgCl2, 0.25 mM dNTP, 0.5 μM of each primer, and 0.5 units Platinum® Taq DNA Polymerase (Invitrogen). Reactions were subjected to PCR amplification: initial incubation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 54 °C for 45 s, and 72 °C for 60 s, and a final incubation at 72 °C for 10 min. PCR products were analyzed on the Bioanalyzer 2100 system. The results were evaluated by comparing every tumor DNA to DNA from the healthy control tissue. Peaks present in the tumor tissue which were not found in the normal tissue indicated instability of a marker. Samples with no altered markers were classified as MSS. Samples with only one altered microsatellite marker were classified as MSI-L. Samples with ≥ 2 altered markers were classified as MSI-H [10].
Statistical analysis
Statistical analysis was done using IBM© SPSS© Statistics version 22 (IBM© Corp., Armonk, NY, USA). Numerical data were expressed as mean and standard deviation or median and range as appropriate. Qualitative data were expressed as frequency and percentage. A chi-square test was used to examine the relation between the qualitative variables. For not normally distributed quantitative data, a comparison between the two groups was done using the Mann-Whitney test (non-parametric t test). All tests were two-tailed. A p value ≤ 0.05 was considered significant.
Results
Patients’ characteristics
Nineteen CRC patients were incorporated in our study, and their clinicopathological features were shown in Table 1. The median age was 40 years (range, 17–72 years), and 8 (42.1%) patients were < 40 years old. CRC was more frequently observed in females (63.2%) than in males (36.8%). Regarding tumor location, 52.6% of the patients had tumor mass in the proximal colon, 31.6% in the distal colon, and only 15.8% in the rectum. Primary tumors were higher in the right colon (52.6%) than in the left side (47.4%). The size of the tumor was observed to be < 5 cm in 14 (73.7%) patients. As regards the TNM staging system, a higher percentage of patients were noted to have tumor stage III (52.6%), followed by stage II (42.1%) and stage IV (5.3%). The N0, N1, and N2 stages of lymph node invasion had percentages of 52.6%, 42.1%, and 5.3%, respectively. The invasion depth of the tumor in the layers of the colon/rectum showed that T3 represented 94.7% of all cases, followed by 5.3% in T4 with no case observed in T1or T2. Finally, the presence of distant metastasis was noticed in only one patient (5.3%).
Somatic mutations results
We determined somatic mutations in 15 genes using targeted NGS on 19 CRC patients, and the mutation list in all patients was described in Table 2. KRAS mutations were detected in 6/19 (31.6%) patients. Codons 12 and 13 constituted 5/6 (83.3%) and 1/6 (16.7%) of all KRAS mutations, respectively. These mutations were substitutions of glycine with aspartate or cyseine on codon 12 and substitution of glycine with aspartate on codon 13. These 3 mutations G12D, G12C, and G13D occur as a result of substitution at c.35G>A, c.34G>T, and c.38G>A and have been detected in 4/6 (66.6%), 1/6 (16.7%), and 1/6 (16.7%) patients, respectively. A higher percentage of KRAS mutations was detected in female patients (5/6, 83.3%) than in male patients (1/6, 16.7%) with no statistically significant difference between the two groups, p value = 0.22. Statistical comparison between CRC patients with wild and mutant KRAS mutation revealed no statistically significant difference between the two groups as regards tumor site, stage, no. of lymph nodes, depth of invasion, and distant metastasis except for their age with p value = 0.003 where mutant KRAS was detected in older CRC patients.
MSI status results
Among the examined 19 CRC patients, 11 (57.9%) patients had MSI-H CRC. A higher percentage of MSI-H CRC was detected in female patients (9/11, 81.8%) than in male patients (2/11, 18.2%) with a statistically significant difference between the two groups, p value = 0.048. Statistical comparison between CRC patients with MSI-H and MSI-L/MSS revealed no statistically significant difference between the two groups as regards their age, tumor site, stage, no. of lymph nodes, depth of invasion, and distant metastasis. Out of 11 MSI-H CRC patients, only 3 patients had mutant KRAS. All mutations were on codon 12 with 2 patients having a substitution of glycine with aspartate and 1 patient having a substitution of glycine with cyseine. Eight patients had both MSI-H CRC and wild KRAS mutation with no statistically significant correlation between MSI status and KRAS mutation in our CRC patients. Correlation of patients' clinicopathological features with KRAS mutations and MSI status was described in Table 3.
Discussion
Colorectal cancer care is becoming increasingly relied on tumor biomarkers to diagnose, select optimal therapy, and anticipate prognosis for each patient. Also, the detection of somatic mutations within patient tumors plays an important role in the risk stratification of CRC patients. Previous studies have shown that KRAS and MSI were prognostic biomarkers for CRC with a significant association of both with the clinicopathological features in those patients [11]. So, the present study aimed to demonstrate the somatic genetic changes in 15 genes especially KRAS mutation using targeted next-generation sequencing and to evaluate MSI status with their correlation to clinicopathological features of Egyptian CRC patients. In our study, KRAS mutation was detected in 31.6% of CRC patients. This is comparatively less than the average level of KRAS mutations (35–45%) found in CRCs, the mutation proportions observed in Serbia, Turkey, Saudi Arabia, and Egypt (32–35.7%), and the mutation proportions observed in Slovenia, Iraq, and Spain (46.2–48%), and higher than the mutation proportions observed in Morocco and Greece and the previously reported proportion in India (23.5–29.3%) [12,13,14]. KRAS mutation was insignificantly higher in female CRC patients, and this is in agreement with our previous study [1]. Other studies found that KRAS mutation was more likely to be noted in male subjects (p = 0.010) [15]. However, in other reports, an even distribution of KRAS mutations has been described in both genders [16]. There was no significant difference in the clinicopathological features between KRAS mutant and KRAS wild patients except for patient age where KRAS mutation was detected in older individuals with p value = 0.003. Other studies found no significant correlation with gender, ethnicity, age, tumor differentiation, histological type, and the presence of distant metastases. However, they found in patients with metastatic disease, KRAS mutations were more common in individuals older than 65 years (p = 0.035) [16]. Regarding MSI status, 57.9% of our patients had MSI-H CRC which was reported as a factor associated with improved prognosis [17]. Previous studies found that MSI-H sporadic CRCs are the most often (about 70–95%) caused by alteration of the MLH1 gene via somatic promoter hypermethylation [18]. In other Egyptian studies, MSI-H was found in 37% and 30.8% of their patients [17, 19]. This difference may be attributed due to different detective methods to some extent. MSI-H CRC was significantly higher in female patients with p value = 0.048. This is in agreement with Tsai et al. who reported that the female patients revealed significantly a MSI-high status (14.0 vs. 8.3%, OR 1.800, p = 0.001) than male patients [20]. However, other reports did not find a significant difference between MSI-H versus MSS/MSI-L regarding gender [21, 22]. Statistical comparison between CRC patients with MSI-H and MSI-L/MSS revealed no statistically significant difference between the two groups as regards their age, tumor site, tumor location, TNM stage, no. of lymph nodes, depth of invasion, and distant metastasis. However, MSI-H CRCs have been reported to demonstrate a more frequent association with proximal tumor location, poorly differentiated, mucin-containing histology, and advanced staging [23]. Another study revealed that proximal location (right side) was selected as predictors of MS-H CRC with p = 0.005 [24]. In our study, only three patients had a mass in the rectum, and they all had MS-H CRC and wild KRAS. However, MSI-H cancers are rarely located in the rectum in Western populations, but fully half of Egyptian MSI-H cases were at that site [19]. Finally, there was no statistically significant correlation between MSI status with KRAS mutation in our CRC patients. However, Ashktorab et al. revealed that the incidence of KRAS mutations is inversely correlated with MSI-H status in CRCs [23]. Again, Liu et al. revealed that KRAS-mutated CRCs had a lower frequency of MSI-H (15% vs. 42%; p = .015) [25].
Conclusion
In conclusion, considering that KRAS mutations confer resistance to EGFR inhibitors, patients who have CRC with KRAS mutation could receive more tailored management by defining MSI status. MSI-high patients have enhanced responsiveness to anti-PD-1 therapies. Thus, the question arises as to whether it is worth investigating this association in the routine clinical setting or not. Further studies with a larger number of patients are needed to assess the impact of MSI status on Egyptian CRC care.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AKT1:
-
AKT serine/threonine kinase 1
- BRAF:
-
B-Raf proto-oncogene, serine/threonine kinase
- CIMP:
-
CpG island methylator phenotype
- CIN:
-
Chromosomal instability
- CRC:
-
Colorectal cancer
- dMMR:
-
MMR-deficient
- EGFR:
-
Epithelial growth factor receptor
- ERBB2:
-
Erb-b2 receptor tyrosine kinase 2
- FOXL2:
-
Forkhead box L2
- GNA11:
-
G protein subunit alpha 11
- GNAQ:
-
G protein subunit alpha q
- IRB:
-
Institutional Review Board
- KIT:
-
KIT proto-oncogene receptor tyrosine kinase
- KRAS:
-
Kirsten rat sarcoma 2 viral oncogene homolog
- MET:
-
MET proto-oncogene, receptor tyrosine kinase
- MMR:
-
Mismatch repair system
- MSI:
-
Microsatellite instability
- MSI-H:
-
Microsatellite instability-high
- MSI-L:
-
Microsatellite instability-low
- MSS:
-
Microsatellite stable
- NGS:
-
Next-generation sequencing
- PDGFRA:
-
Platelet-derived growth factor receptor alpha
- PIK3CA:
-
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- PTEN:
-
Phosphatase and tensin homolog
- RET:
-
Ret proto-oncogene
- STRs:
-
Short tandem repeats
- Tp53:
-
Tumor protein p53
References
Kassem N, Sharaf S, Abdel Aziz A, Mohsen M, Kassem H, El Khateeb S, Medhat N, Nagdy B, Abdel Moneim R, Abdulla M (2018) Towards validation of targeted next-generation sequencing on formalin fixed paraffin embedded colorectal cancer tissues in Egyptian population: a pilot study with feasibility and challenges. Int J Cancer Tremnt 1(1):20–29
Fitzmaurice C, Allen C, Global Burden of Disease Cancer Collaboration et al (2017) Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol 3:524
Hubbard JM, Grothey A (2013) Adolescent and young adult colorectal cancer. J Natl Compr Cancer Netw 11:1219–1225
Worthley DL, Leggett BA (2010) Colorectal cancer: molecular features and clinical opportunities. Clin Biochem Rev 31:31–38
Abd El Kader Y, Emera G, Safwat E, Kassem H, Kassem N (2013) The KRAS StripAssay for detection of KRAS mutation in Egyptian patients with colorectal cancer (CRC): a pilot study. J Egypt Natl Canc Ins 25:37–41
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445
Nojadeh JN, Sharif SB, Sakhinia E (2018) Microsatellite instability in colorectal cancer. EXCLI J 17:159–168 ISSN 1611–2156
Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J (2018) Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 7(3):746–756
Deras IL, Wise A, Zhao C, Glidewell-Kenney C, Le P, Aderhold EU, Gutekunst K (2017) Determination of gene amplifications with a next-generation sequencing cancer panel. In: Proceedings of the American Association for Cancer Research Annual Meeting 2017. AACR; Cancer Res 2017;77(13 Suppl):Abstract nr 3585, Washington, DC. Philadelphia. https://doi.org/10.1158/1538-7445.AM2017-3585
Ortiz C, Dongo-Pflucker K, Martín-Cruz L, Barletta Carrillo C, Mora-Alferez P, Arias A (2016) Microsatellite instability in patients with diagnostic of colorectal cancer. Rev Gastroenterol Peru 36(1):15–22
Hu J, Yan WY, Xie L, Cheng L, Yang M, Li L, Shi J, Liu BR, Qian XP (2016) Coexistence of MSI with KRAS mutation is associated with worse prognosis in colorectal cancer. Medicine 95:50
Bisht S, Ahmad F, Sawaimoon S et al (2014) Molecular spectrum of KRAS, BRAF, and PIK3CA gene mutation: determination of frequency, distribution pattern in Indian colorectal carcinoma. Med Oncol 31(9):1–3
Baskin Y, Calibasi G, Amirfallah A et al (2014) KRAS and BRAF mutation frequencies in a series of Turkish colorectal cancer patients. Transl Cancer Res 3(2):160–166
Malhotra P, Anwar M, Nanda N et al (2013) Alterations in K-ras, APC and p53-multiple genetic pathway in colorectal cancer among Indians. Tumor Biol 34(3):1901–1911
Palomba G, Colombino M, Contu A et al (2012) Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. J Transl Med 10(1):178
Pang XL, Li QX, Ma ZP, Shi Y, Ma YQ, Li XX, Cui WL, Zhang W (2017) Association between clinicopathological features and survival in patients with primary and paired metastatic colorectal cancer and KRAS mutation. Onco Targets Ther 10:2645–2654
Ismael NS, El Sheikh SA, Talaat SM, Salem EM (2017) Mismatch repair proteins and microsatellite instability in colorectal carcinoma (MLH1, MSH2, MSH6 and PMS2): Histopathological and Immunohistochemical Study. Open Access Maced J Med Sci 5(1):9–13
Copija A, Waniczek D, Witkoś A, Walkiewicz K, Nowakowska-Zajdel E (2017) Clinical significance and prognostic relevance of microsatellite instability in sporadic colorectal cancer patients. Int J Mol Sci 18:107
Soliman AS, Bondy ML, El-Badawy SA, Mokhtar N, Eissa S, Bayoumy S, Seifeldin IA, Houlihan PS, Lukish JR, Watanabe T, On On Chan A, Zhu D, Amos CI, Levin B, Hamilton SR (2001) Contrasting molecular pathology of colorectal carcinoma in Egyptian and Western patients. Br J Cancer 85(7):1037–1046
Tsai YJ, Huang SC, Lin HH, Lin CC, Yuan-Tzu Lan YT, Wang HS, Yang SH, Jiang JK, Chen WS, TLin TC, Lin JK, Chang SC (2018) Differences in gene mutations according to gender among patients with colorectal cancer. World J Surg Oncol 16:128
Kang S, Na Y, Joung SY, Lee SL, Oh SC, Min BW (2018) The significance of microsatellite instability in colorectal cancer after controlling for clinicopathological factors. Medicine 97:9
Kim CG, Ahn JB, Jung M, Beom SH, Kim C, Kim JH, Heo SJ, Park HS, Kim JH, Kim NK, Min BS, Kim H, Koom WS, Shin SJ (2016) Effects of microsatellite instability on recurrence patterns and outcomes in colorectal cancers. BJC 115:25–33
Ashktorab H, Ahuja S, Kannan L, Llor X, Ellis NA, Xicola RM, Laiyemo AO, Carethers JM, Brim H, Nouraie M (2016) A meta-analysis of MSI frequency and race in colorectal cancer. Oncotarget 7(23):34546–34557
Soliman NA, Morsi DF, Helmy NAH (2019) Immunohistochemical expression of MMR proteins with clinicopathological correlation in colorectal cancer in Egypt. Open Access Maced J Med Sci 7(10):1608–1617
Liu X, Jakubowski M, Hunt JL (2011) KRAS gene mutation in colorectal cancer is correlated with increased proliferation and spontaneous apoptosis. Am J Clin Pathol 135(2):245–252
Acknowledgements
We deeply acknowledge the scientific and technical support offered by the Oncology Department Molecular lab team. We are also thankful for our dear patients as well as the IT Department team.
Funding
This study was funded by the Kasr Al Ainy Oncology Department, Cairo University, which provides us with the needed kits. This study was performed in the Molecular Lab of Kasr Al-Ainy Oncology Department with the availability of all equipments needed in our study especially MiSeqDX and bioanalyzer.
Author information
Authors and Affiliations
Contributions
NK analyzed and interpreted the patient data. GE provided us with the surgical colonic specimens. MT collected the patients’ samples and was responsible for the molecular testing of the patients samples. NM and BN were responsible for the molecular testing of the patients samples. HK was responsible for the data analysis, bioinformatics and was a major contributor in writing the manuscript. RA was responsible for the demographic and clinical data collection. MA and WM analyzed and interpreted the patient data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Kasr Al Ainy Clinical Oncology Department Institutional Review Board (IRB)-3-2018. Informed consent was obtained from all individual participants included in the study. All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments (GCP guidelines) or comparable ethical standards.
Consent for publication
The consent to publish had been taken from each participant in this work.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kassem, N.M., Emera, G., Kassem, H.A. et al. Clinicopathological features of Egyptian colorectal cancer patients regarding somatic genetic mutations especially in KRAS gene and microsatellite instability status: a pilot study. Egypt J Med Hum Genet 20, 20 (2019). https://doi.org/10.1186/s43042-019-0028-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-019-0028-z