Abstract
Background
The gut microbiota influences host performance playing a relevant role in homeostasis and function of the immune system. The aim of the present work was to identify microbial signatures linked to immunity traits and to characterize the contribution of host-genome and gut microbiota to the immunocompetence in healthy pigs.
Results
To achieve this goal, we undertook a combination of network, mixed model and microbial-wide association studies (MWAS) for 21 immunity traits and the relative abundance of gut bacterial communities in 389 pigs genotyped for 70K SNPs. The heritability (h2; proportion of phenotypic variance explained by the host genetics) and microbiability (m2; proportion of variance explained by the microbial composition) showed similar values for most of the analyzed immunity traits, except for both IgM and IgG in plasma that was dominated by the host genetics, and the haptoglobin in serum which was the trait with larger m2 (0.275) compared to h2 (0.138). Results from the MWAS suggested a polymicrobial nature of the immunocompetence in pigs and revealed associations between pigs gut microbiota composition and 15 of the analyzed traits. The lymphocytes phagocytic capacity (quantified as mean fluorescence) and the total number of monocytes in blood were the traits associated with the largest number of taxa (6 taxa). Among the associations identified by MWAS, 30% were confirmed by an information theory network approach. The strongest confirmed associations were between Fibrobacter and phagocytic capacity of lymphocytes (r = 0.37), followed by correlations between Streptococcus and the percentage of phagocytic lymphocytes (r = -0.34) and between Megasphaera and serum concentration of haptoglobin (r = 0.26). In the interaction network, Streptococcus and percentage of phagocytic lymphocytes were the keystone bacterial and immune-trait, respectively.
Conclusions
Overall, our findings reveal an important connection between gut microbiota composition and immunity traits in pigs, and highlight the need to consider both sources of information, host genome and microbial levels, to accurately characterize immunocompetence in pigs.
Similar content being viewed by others
Background
The pig industry has a considerable socio-economical value representing around 35% of the total meat produced worldwide [1] and being the most popular meat for consumption [2]. The intensification of pig production coupled with the ban on in-feed use of antibiotics has led to a deterioration of the health status of pig farms. In addition, the current emergence of antibiotic resistance and society demands for healthier products and environmentally responsible livestock systems, has motivated to explore relevant approaches for pig and other livestock breeding programs, to improve robustness and disease resistance [3].
The implementation of breeding programs to select animals according to their robustness presents several challenges and levels of complexity. One of the most relevant milestones is the identification of selection criteria that combine functional traits with those of immunocompetence. These complex traits are driven by several physiological and behavioral mechanisms that in turn are determined by genetic and environmental factors. Regarding the genetic determinism of immunocompetence, several studies in pigs acknowledged medium to high heritability estimates [4,5,6,7,8,9] and reported genomic regions and candidate genes associated with phenotypic variation of health-related traits [9,10,11,12,13,14,15].
Over the past few years, multiple studies highlighted the relevant role of the gut microbiota composition in the homeostasis and function of the mammalian immune system [16,17,18,19]. Gut microbiota can regulate host-immunity through both direct mechanisms like translocation of bacteria and their components (i.e., metabolites) or mediate indirect process such as T-cell polarization and the regulation of immune cell trafficking [18]. Commensal gut populations modulate host's immune responses, which in turn can modify the microbiota composition to maintain gut homeostasis [20, 21]. Recently, polymorphisms in immune genes associated with the abundance of microbial communities have been reported [22,23,24,25]. Furthermore, it has been suggested that the pattern recognition receptors, which are proteins capable of recognizing molecules frequently associated with pathogens, may have evolved to mediate the bidirectional crosstalk between microbial symbionts and their hosts [26]. This has resulted in a mutualistic and symbiotic partnership between the immune system and these commensal microorganisms [27]. Therefore, the immune system not only protects the host from pathogens but can also modulate, and is itself modulated by beneficial microbes.
Considering the relevant interplay between gut microbiota and host immunity, a better understanding of the role of gut microbiota in the immunocompetence determination in pigs could greatly assist in the implementation of selection programs to improve robustness and disease resistance simultaneously. The present work aimed to identify microbial biomarkers linked to immunity traits and to estimate the contribution of host-genome and gut microbial communities to immunocompetence in healthy pigs.
Methods
Ethics statement
All experimental procedures were performed according to the Spanish Policy for Animal Protection RD53/2013, which meets the European Union Directive 2010–63-EU about the protection of animals used in experimentation. The experimental protocol was approved by the Ethical Committee of the Institut de Recerca i Tecnologia Agroalimentàries (IRTA).
Animal samples
Samples employed in this study are a subset of pigs reported in Ballester et al. [9]. A total of 389 weaned piglets (196 males and 193 females) from a commercial Duroc pig line were used. The pigs were distributed in six batches obtained from 132 sows and 22 boars. All animals were raised on the same farm and had ad libitum access to the same commercial cereal-based diet.
Immunity and hematological traits
Details of the sampling and laboratory processing have been reported [9]. In brief, blood and saliva samples were collected from all 389 piglets at 60 ± 8 days of age. Blood samples in 4 ml EDTA tubes were used to measure the hemograms (Laboratory Echevarne, Spain; Barcelona). Saliva was collected with Salivette tubes (Sarstedt S.A.U., Germany) according to the protocols recommended by the manufacturer. Blood samples for serum were collected in 6 mL tubes with gel serum separator and centrifuged at 1600 g for 10 min at RT. Plasma was collected from the sampled blood in 6 ml heparinized tubes and centrifuged at 1300 g for 10 min at 4ºC. Plasma and serum samples were collected, aliquoted, and stored a − 80 ºC. The following hematological parameters were included in this study: total number of eosinophils (EO), leukocytes (LEU), lymphocytes (LYM) and neutrophils (NEU) in blood. Analyzed immunity parameters included immunoglobulins (IgA, IgG and IgM) concentrations in plasma; C-reactive protein (CRP), Haptoglobin (HP) and Nitric Oxide (NO) concentrations in serum; and IgA concentration in saliva (IgAsal). Gamma-delta T cells (γδ T cells) were separated from heparinized peripheral blood by density-gradient centrifugation with Histopaque-1077 (Sigma, Spain). Phagocytosis assay was carried out in heparinized whole blood samples incubated with fluorescein (FITC)- labeled opsonized Escherichia coli bacteria using the Phagotest kit (BD Pharmigen, Spain) as indicated in the manufacturer’s protocol. The following phagocytosis traits were used: percentage of total phagocytic cells (PHAGO_%); percentage of phagocytic cells among granulocytes (GRANU_PHAGO_%), monocytes (MON_PHAGO_%) and lymphocytes (LYM_PHAGO_%); mean fluorescence in FITC among the total phagocytic cells (PHAGO_FITC); and mean fluorescence in FITC among the granulocytes (GRANU_ PHAGO_FITC), monocytes (MON_PHAGO_FITC) and lymphocytes (LYM_PHAGO_FITC) that phagocyte.
DNA extraction, sequencing and bioinformatics analysis
Simultaneous with blood and saliva samples, fecal samples were collected from all 389 piglets. DNA was extracted with the DNeasy PowerSoil Kit (QIAGEN, Hilden, Germany) following manufacturer’s instructions. Extracted DNA was sent to the University of Illinois Keck Center for Fluidigm sample preparation and paired-end (2 × 250 nt) sequencing on an Illumina NovaSeq (Illumina, San Diego, CA, USA). The 16S rRNA gene fragment was amplified using the primers V3_F357_N: 5'-CCTACGGGNGGCWGCAG-3' and V4_R805: 5'-GACTACHVGGGTATCTAATCC-3'. Sequences were analysed with Qiime2 [28]; barcode sequences, primers and low-quality reads (Phred scores of < 30) were removed. The quality control also trimmed sequences based on expected amplicon length and removed chimeras. Afterwards, sequences were processed into Amplicon Sequences Variants (ASVs) at 99% of identity. Samples with less than 10,000 reads were excluded and ASVs present in less than three samples and representing less than 0.001% of the total counts were discarded. ASVs were classified to the lowest possible taxonomic level based on a primer-specific trained version of GreenGenes Database [29].
Genotype data
The 389 animals were genotyped using the Porcine 70k GGP Porcine HD Array (Illumina, San Diego, CA) containing 68,516 single nucleotide polymorphisms (SNPs). The quality control excluded SNPs with minor allele frequencies < 5%, rates of missing genotypes above 10%, and SNPs that did not map to the porcine reference genome (Sscrofa11.1 assembly). Consequently, 42,641 SNPs were retained for subsequent analysis.
Microbiability and heritability estimation
Heritability (h2), i.e. the proportion of variance explained by the host genetics, and microbiability (m2), i.e. the proportion of variance explained by the microbial composition, were estimated for each immunity trait based on a mixed-model as follows:
where y is the n-dimensional vector containing the individual phenotypes for the immune trait under consideration; β is the vector of fixed effects, containing the general intercept, the sex effect (two levels), and batch effect (six levels) for most traits but data of laboratory analysis (12 levels, two by batch) for phagocytosis-related traits; u is the vector containing the host genetic random effect from each individual; m is the vector of the animal’s microbiome random effect; X, Z and W are, respectively, the incidence matrices correspondent to β, u and m; and e is the vector of residual terms.
Assuming independence between random effects, the following distributions were considered: u ~ N(0,G, σ2u), where σ2u is the host genetic effects variance and G is the genomic relationship matrix between individuals, computed following [30], i.e., \(G = \frac{{ss^{{\prime }} }}{{2\sum\nolimits_{i} {p_{i} (1 - p_{i} )} }}\) being S the matrix that contains the centered individual genotype for the 42,641 SNPs (columns) of each individual (rows), and pj is the frequency of the minimum allele of the jth SNP; m ~ N(0,B, σ2m), where σ2m is the microbial effects variance and B the microbial relationship matrix computed following [31], i.e., \(B = \frac{{MM^{{\prime }} }}{n}\), being M the matrix containing the scaled after a previous cumulative sum scaling normalization of the ASV abundances (columns) for each individual microbiome (rows) and n the total number of ASVs; and finally \(e \sim N(0,I\sigma_{e}^{2} )\), where \(\sigma_{e}^{2}\) is the error variance.
The model parameters for each immunity trait were estimated by a Bayesian approach, using the Bayes Ridge Regression model from BGLR package [32]. We used a Gibbs sampler with 30,000 iterations and a burn-in of 3,000 rounds. The ‘heritability’ \(\left( {h^{2} = \frac{{\sigma_{u}^{2} }}{{\left( {\sigma_{u}^{2} + \sigma_{m}^{2} + \sigma_{e}^{2} } \right)}}} \right)\) and ‘microbiability’ \(\left( {m^{2} = \frac{{\sigma_{m}^{2} }}{{\left( {\sigma_{u}^{2} + \sigma_{m}^{2} + \sigma_{e}^{2} } \right)}}} \right)\) were estimated from the mean of the posterior distributions [33].
Microbial wide association study
We performed a Microbial Wide Association Study (MWAS) using a multi-ASV association method that combines all the ASVs in a single model:
Given a trait yi measured in n individuals and a matrix X containing relative abundances of p taxa from a microbial community, here the ASVs effects were treated as draws from normal distributions as in any Bayesian Ridge Regression approach [32].
Following the approach of Legarra et al. [34], Bayes Factor (BF) for the effect of each taxa can be derived as the ratio of probabilities \(BF = \frac{{P_{H1} (y)}}{{P_{H0} (y)}}\), where H1 means “the j-genus has some effect” and H0 “the j-genus has no effect”. The calculations from the posterior distribution are very simple since both probabilities \((P_{H1} ,P_{H0} )\) are normal density.
Network between microbial and immunity traits
To better understand the relationship between microbial communities and immunity traits we implemented PCIT [35], a network-based approach that combines partial correlation coefficient with information theory to identify significant correlations between each possible combination of clr-transformed bacterial abundance and the immune-traits [35]. The PCIT algorithm is a data-drive semi-parametric approach that does not require independence to make the results interpretable. Instead, it uses the definition of the partial correlation between “x” and “y” given “z” as being the strength of the linear relationship between “x” and “y” that is independent from “z”. In doing so, PCIT tests all possible 3-way combinations in the dataset and only keeps correlations between traits if they are significant and independent of the association of another features. To reduce the complexity of the resulting network, from the PCIT significant connections, we kept only the ones involving one immune-trait and one genus (i.e. genus-genus and trait-trait interactions were no represented).
Results
In this study, 16S rRNA gene sequences, host genotype information and immune traits from 389 Duroc pigs were analyzed to estimate both host genomes and gut microbiota contribution to the porcine immunocompetence, and to identify microbial biomarkers linked to immunity traits. Table 1 summarizes the immunity traits and their descriptive statistics used in the present study. Regarding 16S rRNA gene sequences, after quality control, a total of 2,055 Amplicon Sequences Variants (ASVs) and 68 genera were detected. The dominant bacterial phyla were Bacteroidetes and Firmicutes, and the most abundant genera were Prevotella, Lactobacillus, Treponema, Roseburia and Ruminococcus (Additional file 1: Fig. 1).
Heritability and microbiability of immunity traits
Posterior estimates of h2 and m2 for the 21 health-related traits can be shown in Fig. 1 and Additional file 2: Table 1. Posterior means of h2 in the analyses considering microbiota contribution reached low to medium values (from 0.138 to 0.359), but posterior probability of h2 being superior to 0.1 was in all cases above 0.82. Similarly, estimated m2 reached values between 0.152 and 0.276, and the probability of being above 0.1 was above 0.85 for all immunity and hematological traits (Additional file 2: Table 1). Among analysed traits, IgG and IgM in plasma showed the highest genetic determinism (h2 = 0.316 and 0.359), whereas microbiota contribution was below 0.18. Conversely, the Hp concentration in serum showed the highest microbial effect (m2 = 0.276), accompanied by the lowest h2 estimate (h2 = 0.138). Considering the joint effects of host-genome and gut microbiota, these two sources of variation explained from 29.9% to 51.7% phenotypic variance of the analysed immunity and hematological traits. To be noted, in the 76% (16/21) of these traits the h2 and m2 estimates reaching relatively similar values (Fig. 1).
Associations between microbial genera and immunity traits
Results from the MWAS reported some putative associations between bacterial genera abundance and health-related traits (Table 2). In particular, 15 out of the 21 immunity traits were associated with at least one microbial genus (Table 2). The strongest association was observed between the relative abundance of Chlamydia and the profile of LYM_PHAGO_FITC, followed by Streptococcus linked to LYM_PHAGO_% and Peptococcus associated with LYM_PHAGO_FITC. In addition, several genera showed multiple associations with numerous immunity traits: Desulfovibrio, Oribacterium and Chlamydia (4 traits) followed by Oxalobacter and Parabacteroides (3 traits), Peptococcus and Streptococcus (2 traits). As far as the analysed phenotypes, those traits showing the highest number of associations with different bacterial taxa were: LYM_PHAGO_FITC and MON (6 taxa); LYM_PHAGO_%, EOS, GRANU_PHAGO_FITC (4 taxa) and total number of LEU (3 taxa). Meanwhile, only four out of the 15 immunity traits analysed were linked with only one genus (Table 2).
Gut microbial and host-immune interaction network
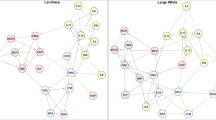
The interplay between microbial and health-related traits was also inferred through a network comprised of 63 nodes (42 genera and 21 immunity traits) and 86 edges (significant connections) in which only the significant interactions between a bacterial genus and an immunity trait were considered (Fig. 2). The topological evaluation of the network highlights LYM_PHAGO_% as the most connected trait, followed by IgAsal, NEU and Hp. Meanwhile, at microbial level, Streptococcus was the most connected genus followed by Acidaminococcus, Desulfovibrio and Blautia. The network approach confirmed 30% (12/40) of the associations identified by the MWAS (Fig. 2). The strongest confirmed correlation was between Fibrobacter and LYM_PHAGO_FITC (r = 0.37) followed by correlations between Streptococcus and LYM_PHAGO_% (r = -0.34) and between Megasphaera and Hp (r = 0.26). To be noted, Streptococcus and LYM_PHAGO_% that showed the strongest confirmed association in the MWAS were highly in the interaction-network as the keystone bacterial and immunity trait, respectively.
Microbial—health-related traits network. Green diamond nodes correspond to immunity traits (n = 21) and blue ellipse nodes correspond to microbial genera (n = 42). Node sizes are relative to their topological degree (number of connections) and edges are continuous or dashed to represent positive or negative correlations, respectively. Relationships previously identified by MWAS are highlighted in red
Discussions
Host-genome and gut microbiota contribution to porcine immunocompetence
We report the first study that aimed to dissect the joint contribution of the host genome and the gut microbiota to the immunocompetence in healthy pigs. Estimates of microbiability pointed out significant microbial effects on most immunity and hematological traits, ranging between 15 and 27% of total phenotypic variance. Effects of microbiota resulted particularly relevant for Hp concentrations in serum, followed by the parameters related to phagocytosis of lymphocytes. Regarding genomic heritabilities of these traits, they reached low to moderate values and were substantially lower compared to the medium to high h2 previously obtained in the same Duroc population [9] for all traits but MON and MON_PHAGO_%. A dramatic decrease of the estimated host genetic effects was observed for γδ T cells, but also for EOS and NEU counts and immunoglobulins concentrations in plasma, despite IgM and IgG variability seemed dominated by host genetics and showed the highest h2 among analysed traits. These results would call into question the high genetic determinism of the global immunocompetence in pigs reported in previous studies [6, 7, 9]. However, it should be considered that the limited sample size joint with the likely similarity between close relatives (particularly between full-sibs) in their microbiota profiles makes plausible that the model could not separate adequately genetic from microbiota effects.
Microbial signatures associated with immunity traits
In the present study, we implemented a combination of MWAS and network approaches to pinpoint microbial signatures associated with immunity traits, revealing some interesting associations between the composition of the pig gut microbiota and the host immunity traits. Remarkably, lymphocyte phagocytosis traits were among the most connected and associated traits to the highest number of taxa and were also central nodes in the network. The strongest confirmed association involved Fibrobacter relative abundance in gut microbiota and the host phagocytosis capacity of lymphocytes, which were positively correlated (r = 0.37). Fibrobacter genus is composed of strictly anaerobic bacteria with cellulolytic capacity capable of degrading complex plant fiber [36] and it has been associated with better feed efficiency in pigs [37, 38]. Conversely, the relative abundance of Streptococcus showed an opposite association with the percentage of phagocytic lymphocytes (r = -0.34). Streptococcus was also the keystone taxa in the network. In pigs, some Streptococcus species are important opportunistic pathogens such as Streptococcus suis, which abundance increased in the stomach and small intestine after weaning [39]. Piglets with high intestinal concentrations of S. suis can serve as a source of transmission and infection between animals and farms (reviewed in [39]). In general Streptococcus are less abundant in more-feed efficient pigs [37], although there are also evidences of the immunomodulatory properties of members of Streptococcus genus, such as Streptococcus thermophilus, considered beneficial for the organism [40,41,42].
Several studies in mammals have demonstrated that B cells have a significant phagocytic capacity, being able to phagocytose particles including bacteria [43,44,45]. Most important has been the demonstration of the efficient capability of these cells to present antigen from phagocytosed particles to CD4+ T cells [43,44,45], acting as a bridge that link innate with adaptive immunity. Therefore, considering the inferred high connection of these phagocytosis phenotypes with gut microbiota, we could hypothesize that, as other antigen-presenting cells such as dendritic cells or macrophages, the phagocytic lymphocytes seem to be relevant to maintain immune tolerance to the normal gut microbiota, being also relevant to control the abundance of opportunistic pathogens. B-cells also produce secretory IgA, the most abundant secreted isotype in mammals and a key element to maintain ‘homeostatic immunity’[46]. Secretory IgA was among the most connected traits in the network being positively associated with several taxa. Among them, the genus Blautia is of particular interest due to its potential role modulating inflammatory and metabolic diseases, with potential beneficial effects for the host [47]. Therefore, similar to phagocytic lymphocytes, the interplay between secretory IgA levels and the abundance of different taxa in our animals may regulate the ecological balance of commensal bacteria and the development of Ig-A secreting cells. Neutrophils were also positively correlated with gut microbiota profiles. A systemic immunomodulation of neutrophils by intestinal microbiota has been demonstrated [48], and a crosstalk between NEU and gut microbial composition has been also documented [49]. Our results confirmed a positive association of Oribacterium abundance with the quantity of neutrophils. Oribacterium genus belongs to the Lachnospiraceae family, and the abundance of this genus increased in piglets after weaning [50]. Members of the genus Oribacterium produce short-chain fatty acids such as acetate [51], which directly influences immune system regulation [52], and can contribute to the health of the pig.
Among the most connected traits in our network we also found acute-phase protein Hp, which based on the estimated microbiability appeared preferentially determined by microbiota effects. The main function of Hp is to facilitate hemoglobin (Hb) clearance. After the formation of stable Hp-Hb complexes, the macrophage receptor CD163 recognize them and the entire complex is removed from circulation by receptor-mediated endocytosis [53]. Therefore, Hp favors the reduction of free iron concentrations in the circulation and tissues [54]. Several bacterial pathogens such as Staphylococcus, Mycobacteria, Salmonella, Corynebacterium, Haemophilus, among others, require iron for growth, thus elaborating different acquisition strategies to uptake heme from the host, particularly from Hb [55,56,57]. The host immune system has developed antimicrobial mechanisms, most related to innate pathways, to deplete iron availability for pathogens [55]. Remarkably, our results indicate a relevant effect of the microbiota composition on Hp levels which could also modulate the concentration of circulating free iron. We could hypothesize that the symbiotic microbiota could also modulate the iron levels in these animals through innate immunity mechanisms to prevent the development of different opportunistic pathogens. Our results confirmed a positive association between serum concentration of Hp and the relative abundance of Megasphaera, a member of the phylum Firmicutes. According to this result, an increase in the abundance of Megasphaera has been described in colon content and faeces of pigs fed with iron-deficient diet [58]. Interestingly, this genus was reported as a potential biomarker for immune-mediate mechanism of protection from diarrhea [59] and positive correlated with luminal IgA concentration in pigs [50].
Finally, it is worth highlighting the negative association between the relative abundance of Desulfovibrio and LEU and MON counts. Desulfovibrio is a sulfate-reducing bacteria (SRB), which can promote the metabolism of sugars [60] and plays also a key role in intestinal hydrogen and sulfur metabolism [61]. In pigs, Desulfovibrio plays a relevant role during pig gut colonization [50] and was among the dominant genus in healthy pigs compared with diarrhea-affected piglets [62]. In fact, in weaned piglets, a negative correlation between Desulfovibrio and several inflammatory markers such as IL-1β, IL-2 and IL-6, have been observed [63], which would be in agreement with the negative correlation observed between Desulfovibrio and LEU and MON counts in our piglets.
Despite of an inventory of potential gut health biomarkers exists for pigs [64, 65], our results propose new microbial candidates, and emphasize a polymicrobial nature of the immunocompetence in pigs. Furthermore, in agreement with previous reports [66], our results suggest that some immunity traits are influenced by specific microorganisms while others are determined by interactions between members of the gut microbiome. We are aware of some limitations of our study, such as the narrow taxonomic resolution achieved by targeting the V3-V4 16S rRNA variable region with short-read sequencing platforms. Additionally, other sources of variation not controlled in our study at both microbial (e.g. microbial-derived metabolites) and host levels could have been playing an immunomodulatory role. It would have also been desirable counting on early-life microbial records, as this is a critical developmental stage in the maturation of the host immune system, the development of long-term homeostatic immunity and disease susceptibility in the adulthood [67, 68].
In summary, we reveal the joint contribution of the host genome and the gut microbial ecosystem to the phenotypic variance of immunity parameters, and advice that ignoring microbial effects could generate an overestimation of genetic parameters. Further exploration of the mechanisms harmonizing the host and microbial contribution to homeostatic immunity will allow developing holistic breeding strategies to modulate immunocompetence, as well as to improve animal health, robustness and welfare.
Conclusions
Estimates of heritability and microbiability exposed the joint contribution of both the host genome and the gut microbial ecosystem to the phenotypic variance of immunity parameters, and revealed that ignoring microbiota effects on phenotypes could generate an upward bias in the estimation of genetic parameters. Results from the MWAS suggested a polymicrobial nature of the immunocompetence in pigs and highlighted associations between the compositions of pig gut microbiota and 15 of the analyzed traits. Overall, our findings establish several links between the gut microbiota and the immune system in pigs, underscoring the importance of considering both sources of information, host-genome and microbial level, for the genetic evaluation and the modulation of immunocompetence in pigs.
Availability of data and materials
The raw sequencing data employed in this article has been submitted to the NCBI’s sequence read archive (https://www.ncbi.nlm.nih.gov/sra); BioProject: PRJNA608629.
Abbreviations
- CRP:
-
C-reactive protein in serum
- EO:
-
Eosinophils count
- γδ T-cells:
-
γδ T-lymphocyte subpopulation
- GRANU_PHAGO_FITC:
-
Granulocytes phagocytosis
- GRANU_PHAGO_%:
-
Granulocytes phagocytosis
- HP:
-
Haptoglobin in serum
- IgA:
-
IgA in plasma
- IgAsal:
-
IgA in saliva
- IgG:
-
IgG in plasma
- IgM:
-
IgM in plasma
- LEU:
-
Leukocytes count
- LYM:
-
Lymphocytes count
- LYM_PHAGO_FITC:
-
Lymphocytes phagocytosis FITC
- LYM_PHAGO_%:
-
Lymphocytes phagocytosis
- MON_PHAGO_FITC:
-
Monocytes phagocytosis FITC
- MON_PHAGO_%:
-
Monocytes phagocytosis
- MON:
-
Monocytes count
- NO:
-
Nitric oxide in serum
- PHAGO_FITC:
-
Phagocytosis FITC
- PHAGO_%:
-
Phagocytosis (% cells)
- NEU:
-
Neutrophils count
- QIIME:
-
Quantitative insights into microbial ecology
- clr:
-
Centered log ratio transformation
- MWAS:
-
Microbial-wide association studies
- PCIT:
-
Partial Correlation coefficient with Information Theory
References
OECD, Food, Nations AOotU: OECD-FAO agricultural outlook 2019–2028; 2019.
Briefs EAM: World food consumption patterns—trends and drivers. http://ec.europa.eu/agriculture/markets-and-prices/market-briefs/index_en.htm; 2015.
A Reverter BC Hine L Porto-Neto Y Li CJ Duff S Dominik AB Ingham 2021 ImmuneDEX: a strategy for the genetic improvement of immune competence in Australian Angus cattle LID https://doi.org/10.1093/jas/skaa384 1525-3163 (Electronic)
AH V, LL J, TA N, KH Dg. Disease incidence and immunological traits for the selection of healthy pigs. A. D - 7909485 (- 0165–2176 (Print)):- 29–34.
I E-L, E W, U M, C F. Genetic variation in parameters reflecting immune competence of swine. D - 8002006 (- 0165–2427 (Print)):- 1–16.
M Clapperton AB Diack O Matika EJ Glass CD Gladney MA Mellencamp A Hoste SC Bishop 2009 Traits associated with innate and adaptive immunity in pigs: heritability and associations with performance under different health status conditions Genet Sel Evol 41 1 54 54
L F, Y G, D L, G L, JJ L, A T, AM C, J L, P P, C dV et al. - Immunity traits in pigs: substantial genetic variation and limited covariation. D - 101285081 (- 1932–6203 (Electronic)):- e22717.
I E-L, E W, L M, M M, L A-E, L A, C F. Mapping quantitative trait loci for immune capacity in the pig. D - 2985117r (- 0022–1767 (Print)):- 829–835.
M Ballester Y Ramayo-Caldas O González-Rodríguez M Pascual J Reixach M Díaz F Blanc S López-Serrano J Tibau R Quintanilla 2020 Genetic parameters and associated genomic regions for global immunocompetence and other health-related traits in pigs Sci Rep 10 1 1 15
W Luo S Chen D Cheng L Wang Y Li X Ma X Song X Liu W Li J Liang 2012 Genome-wide association study of porcine hematological parameters in a large white× Minzhu F2 resource population Int J Biol Sci 8 6 870 881
S Bovo G Mazzoni F Bertolini G Schiavo G Galimberti M Gallo S Dall’Olio L Fontanesi 2019 Genome-wide association studies for 30 haematological and blood clinical-biochemical traits in Large White pigs reveal genomic regions affecting intermediate phenotypes Sci Rep 9 1 7003
Wang JY, Luo Yr Fau - Fu WX, Fu Wx Fau - Lu X, Lu X Fau - Zhou JP, Zhou Jp Fau - Ding XD, Ding Xd Fau - Liu JF, Liu Jf Fau - Zhang Q, Zhang Q. Genome-wide association studies for hematological traits in swine. (1365–2052 (Electronic)).
Jung EJ, Park HF, Lee JB, Lee JF, Yoo CK, Yoo CF, Kim BM, Kim BF, Kim HI, Kim HF, Cho IC, Cho IF, Lim HT, Lim HT. Genome-wide association study identifies quantitative trait loci affecting hematological traits in an F2 intercross between Landrace and Korean native pigs. (1365–2052 (Electronic)).
Ponsuksili S, Reyer H, Trakooljul N, Murani E, Wimmers KA-O. Single- and Bayesian multi-marker genome-wide association for haematological parameters in pigs. (1932–6203 (Electronic)).
Yan G, Guo T, Xiao S, Zhang F, Xin W, Huang T, Xu W, Li Y, Zhang Z, Huang L. Imputation-based whole-genome sequence association study reveals constant and novel loci for hematological traits in a large-scale Swine F(2) resource population. (1664–8021 (Print)).
P Vernocchi F Chierico Del L Putignani 2016 Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health Front Microbiol 7 1144
J Schluter JU Peled BP Taylor KA Markey M Smith Y Taur R Niehus A Staffas A Dai E Fontana 2020 The gut microbiota is associated with immune cell dynamics in humans Nature 588 7837 303 307
BC Lo GY Chen G Núñez R Caruso 2021 Gut microbiota and systemic immunity in health and disease Int Immunol 33 4 197 209
Estelle J, Mach N, Ramayo-Caldas Y, Levenez F, Lemonnier G, Denis C, Doré J, Larzul C, Lepage P, Rogel-Gaillard C. The influence of host’s genetics on the gut microbiota composition in pigs and its links with immunity traits. In: 10th World Congress of Genetics Applied to Livestock Production. Vancouver, BC, Canada; 2014.
Kamada N, Seo Su Fau - Chen GY, Chen Gy Fau - Núñez G, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. (1474–1741 (Electronic)).
NM Gerardo KL Hoang KS Stoy 1808 Evolution of animal immunity in the light of beneficial symbioses Philos Trans R Soc B Biol Sci 2020 375 20190601
AA Khan L Yurkovetskiy K O’Grady JM Pickard R Pooter de DA Antonopoulos T Golovkina A Chervonsky 2019 Polymorphic immune mechanisms regulate commensal repertoire Cell Rep 29 3 541 550.e544
J W, Id O, L C, N Z, X X, Y X, B Z: Of genes and microbes: solving the intricacies in host genomes. D - 101532368 (1674–8018 (Electronic)): 446–461.
Ramayo-Caldas Y, Prenafeta-Boldú F, Zingaretti LM, Gonzalez-Rodriguez O, Dalmau A, Quintanilla R, Ballester M: Gut eukaryotic communities in pigs: diversity, composition and host genetics contribution. Anim Microbiome 2020, 2(1), 1-12.
A Reverter M Ballester PA Alexandre E Mármol-Sánchez A Dalmau R Quintanilla Y Ramayo-Caldas 2021 A gene co-association network regulating gut microbial communities in a Duroc pig population Microbiome 9 1 52
H C, SK M: - Innate immune recognition of the microbiota promotes host-microbial symbiosis. D - 100941354 (1529–2916 (Electronic)): 668–675.
F B, RE L, JL S, DA P, JI G. Host-bacterial mutualism in the human intestine. D - 0404511 (1095–9203 (Electronic)): 1915–1920.
E Bolyen JR Rideout MR Dillon NA Bokulich CC Abnet GA Al-Ghalith H Alexander EJ Alm M Arumugam F Asnicar 2019 Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 Nat Biotechnol 37 8 852 857
TZ DeSantis P Hugenholtz N Larsen M Rojas EL Brodie K Keller T Huber D Dalevi P Hu GL Andersen 2006 Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB Appl Environ Microbiol 72 7 5069 5072
PM VanRaden 2008 Efficient methods to compute genomic predictions J Dairy Sci 91 11 4414 4423
GF Difford DR Plichta P Løvendahl J Lassen SJ Noel O Højberg A-DG Wright Z Zhu L Kristensen HB Nielsen 2018 Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows PLoS Genet 14 10 e1007580
P Pérez G Campos de los 2014 Genome-wide regression and prediction with the BGLR statistical package Genetics 198 2 483 495
G Campos de los D Sorensen D Gianola 2015 Genomic heritability: what is it? PLOS Genet 11 5 e1005048
A Legarra A Ricard L Varona 2018 GWAS by GBLUP single and multimarker EMMAX and Bayes factors with an example in detection of a major gene for horse gait G3 Genes Genomes Genet 8 7 2301
Reverter A, Chan EK. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. (1367–4811 (Electronic)).
AP Neumann CA McCormick G Suen 2017 Fibrobacter communities in the gastrointestinal tracts of diverse hindgut-fermenting herbivores are distinct from those of the rumen Environ Microbiol 19 9 3768 3783
GE Gardiner BU Metzler-Zebeli PG Lawlor 2020 Impact of intestinal microbiota on growth and feed efficiency in pigs: a review Microorganisms 8 12 1886
McCormack UM, Curião T, Metzler-Zebeli BU, Wilkinson T, Reyer H, Crispie F, Cotter PD, Creevey CJ, Gardiner GE, Lawlor PG. Seeking to improve feed efficiency in pigs through microbial modulation via fecal microbiota transplantation in sows and dietary supplementation of offspring with inulin. Appl Environ Microbiol 2019;AEM.01255–01219.
Y Su W Yao ON Perez-Gutierrez H Smidt W-Y Zhu 2008 Changes in abundance of Lactobacillus spp. and Streptococcus suis in the stomach, jejunum and ileum of piglets after weaning FEMS Microbiol Ecol 66 3 546 555
B Bogert van den M Meijerink EG Zoetendal JM Wells M Kleerebezem 2014 Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota PLoS ONE 9 12 e114277
Perdigon GF, Nader de Macias ME, Nader de Macias MF, Alvarez S, Alvarez SF, Oliver G, Oliver GF, Pesce de Ruiz Holgado AA, Pesce de Ruiz Holgado AA. Enhancement of immune response in mice fed with Streptococcus thermophilus and Lactobacillus acidophilus. (0022–0302 (Print)).
H Mizuno K Tomotsune MA Islam R Funabashi L Albarracin W Ikeda-Ohtsubo H Aso H Takahashi K Kimura J Villena 2020 Exopolysaccharides from Streptococcus thermophilus ST538 modulate the antiviral innate immune response in porcine intestinal epitheliocytes Front Microbiol 11 894
D Parra AM Rieger J Li Y-A Zhang LM Randall CA Hunter DR Barreda JO Sunyer 2012 Pivotal advance: peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+ T cells J Leukoc Biol 91 4 525 536
J Gao X Ma W Gu M Fu J An Y Xing T Gao W Li Y Liu 2012 Novel functions of murine B1 cells: active phagocytic and microbicidal abilities Eur J Immunol 42 4 982 992
A Martínez-Riaño ER Bovolenta P Mendoza CL Oeste MJ Martín-Bermejo P Bovolenta M Turner N Martínez-Martín B Alarcón 2018 Antigen phagocytosis by B cells is required for a potent humoral response EMBO Rep 19 9 e46016
Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. 2017(1097–4180 (Electronic)).
X Liu B Mao J Gu J Wu S Cui G Wang J Zhao H Zhang W Chen 2021 Blautia—a new functional genus with potential probiotic properties? Gut Microbes 13 1 1 21
H-J Wu E Wu 2012 The role of gut microbiota in immune homeostasis and autoimmunity Gut Microbes 3 1 4 14
D Zhang PS Frenette 2019 Cross talk between neutrophils and the microbiota Blood 133 20 2168 2177
N Mach M Berri J Estellé F Levenez G Lemonnier C Denis J-J Leplat C Chevaleyre Y Billon J Doré 2015 Early-life establishment of the swine gut microbiome and impact on host phenotypes Environ Microbiol Rep 7 3 554 569
MV Sizova PA Muller D Stancyk NS Panikov M Mandalakis A Hazen T Hohmann SN Doerfert W Fowle AM Earl 2014 Oribacterium parvum sp. nov. and Oribacterium asaccharolyticum sp. nov., obligately anaerobic bacteria from the human oral cavity, and emended description of the genus Oribacterium Int J Syst Evolut Microbiol 64 8 2642 2649
Smith PM, Howitt MF, Panikov N, Panikov NF, Michaud M, Michaud MF, Gallini CA, Gallini CF, Bohlooly YM, BohloolyYMF, Glickman JN, Glickman JF, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. 2013(1095–9203 (Electronic)).
MJ N, CB A, SK M. CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point. D - 2985121r (1083-351X (Electronic)): 18834–18841.
M MacKellar DJ Vigerust 2016 Role of haptoglobin in health and disease: a focus on diabetes Clin Diabetes 34 3 148 157
EE Johnson M Wessling-Resnick 2012 Iron metabolism and the innate immune response to infection Microbes Infect 14 3 207 216
JE Choby EP Skaar 2016 Heme Synthesis and Acquisition in Bacterial Pathogens J Mol Biol 428 17 3408 3428
TW S, DJ M, PW W, R W, SD K, TM V, TL S: - Complex role of hemoglobin and hemoglobin-haptoglobin binding proteins in. D - 0246127 (0019-9567 (Print)): 6213–6225.
LC Knight M Wang SM Donovan RN Dilger 2019 Early-life iron deficiency and subsequent repletion alters development of the colonic microbiota in the pig Front Nutr 6 120
Carey MA, Medlock GL, Alam M, Kabir M, Uddin MJ, Nayak U, Papin J, Faruque ASG, Haque R, Petri WA, Jr., et al. Megasphaera in the stool microbiota is negatively associated with diarrheal cryptosporidiosis. Clin Infect Dis. 2021.
N Salazar M Gueimonde AM Hernández-Barranco P Ruas-Madiedo CG Reyes-Gavilán de 2008 Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria Appl Environ Microbiol 74 15 4737
S Ran C Mu W Zhu 2019 Diversity and community pattern of sulfate-reducing bacteria in piglet gut J Anim Sci Biotechnol 10 1 40
E Ramon L Belanche-Muñoz F Molist R Quintanilla M Perez-Enciso Y Ramayo-Caldas 2021 kernInt: a kernel framework for integrating supervised and unsupervised analyses in spatio-temporal metagenomic datasets Front Microbiol 12 60
R Hu Z He M Liu J Tan H Zhang D-X Hou J He S Wu 2020 Dietary protocatechuic acid ameliorates inflammation and up-regulates intestinal tight junction proteins by modulating gut microbiota in LPS-challenged piglets J Anim Sci Biotechnol 11 1 1 12
Wijtten PJ, van der Meulen J Fau - Verstegen MWA, Verstegen MW: Intestinal barrier function and absorption in pigs after weaning: a review. (1475–2662 (Electronic)).
N Theo Eds 2014 Intestinal health Wageningen Academic Publishers
AC Flamme La S Milling 2020 Immunological partners: the gut microbiome in homeostasis and disease Immunology 161 1 1 3
E Ansaldo TK Farley Y Belkaid 2021 Control of immunity by the microbiota Annu Rev Immunol 39 1 449 479
D Zheng T Liwinski E Elinav 2020 Interaction between microbiota and immunity in health and disease Cell Res 30 6 492 506
Acknowledgements
The authors warmly thank all technical staff from Selección Batallé S.A, for providing the animal material and their collaboration during the sampling.
Funding
YRC is recipient of a Ramon y Cajal post-doctoral fellowship (RYC2019-027244-I) from the Spanish Ministry of Science and Innovation. MB was recipient of a Ramon y Cajal post-doctoral fellowship (RYC-2013–12573). LMZ is recipient of a Ph.D. grant from Ministry of Economy and Science, Spain associated with ‘Centro de Excelencia Severo Ochoa 2016–2019’ award SEV-2015-0533 to CRAG. Part of the research presented in this publication was funded by Grants AGL2016-75432-R, AGL2017-88849-R awarded by the Spanish Ministry of Economy and Competitiveness. The authors belong to Consolidated Research Group TERRA (AGAUR, 2017 SGR 1719).
Author information
Authors and Affiliations
Contributions
YRC, RQ and MB designed the study. MB carried out DNA extraction. MB, RQ, TD and YRC performed the sampling. YRC, LMZ, AR and PA analyzed the data. YRC, LMZ, DP, AR, PA, RQ and MB interpreted the results and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Animal care and experimental procedures were carried out following national and institutional guidelines for the Good Experimental Practices and were approved by the IRTA Ethical Committee. Consent to participate is not applicable in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Figure 1. Iris-plot representing the 20 most abundant genera. Each bar represents a sample, and bar colors represented the genera relative abundance.
Additional file 2.
Table 1. Posterior estimates of h2 and m2 for the 21 health-related traits.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ramayo-Caldas, Y., Zingaretti, L.M., Pérez-Pascual, D. et al. Leveraging host-genetics and gut microbiota to determine immunocompetence in pigs. anim microbiome 3, 74 (2021). https://doi.org/10.1186/s42523-021-00138-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42523-021-00138-9