Abstract
Background
β-catenin is an important unit of the Wnt/β-catenin signaling pathway, a conserved process involving several physiological activities, encompassing differentiation and cell proliferation, etc. The dysfunction or mutation in β-catenin causing the initiation and advancement of various neoplasm types, including colorectal cancer, breast cancer, etc., has been reported. Therefore, β-catenin is a therapeutic target. Hence, designing new inhibitors targeted against β-catenin will prevent cancerous cells’ involvement and eliminate the diseases. Studies showed that Vitis vinifera, a well-known grape species, contains different phytochemical substances, including aromatic acids, flavonoids, phenolic compounds, proanthocyanins, etc. V.vinifera exerts different anticancer properties such as apoptosis, cell proliferation, cell cycle arrest, and inhibition in cancerous cells. Structural bioinformatics methods, including molecular docking, molecular mechanics generalized Born surface area (MM/GBSA), absorption, distribution, metabolism, excretion studies (ADMET), and pharmacophore modeling approach, were used to determine the potential β-catenin inhibitors from V.vinifera bioactive compounds.
Result
Cis-astringin, rutin, caftaric acid, trans-caftaric acid, procyanidin B3, cis-Miyabenol C, and ampelopsin H are shown to be suitable inhibitors against β-catenin due to their binding affinity and interaction with the amino acids residues at the binding sites of β-catenin compared to Food and Drug Administration (FDA) approved drugs leucovorin Calcium and Xeloda prescribed to cure colorectal cancer.
Conclusions
This study suggests that V. vinifera could be a good plant source for compounds that might treat cancer by inhibiting the Wnt/β-catenin signaling pathway.
Similar content being viewed by others
Background
The Wnt/β-catenin pathway, otherwise named the canonical Wnt pathway, is a preserved mechanism that participates in different physiological activities, including apoptosis, cell proliferation, tissue homeostasis, and differentiation (Choi et al. 2020; Salik et al. 2020; Soleas et al. 2020). The dysfunction of β-catenin, a significant part of the Wnt signaling pathway, has been connected with tumors’ initiation and progression. High level of β-catenin occurs when there is overexpression of the canonical WNT pathway. Hence, mutations in the Wnt/β-catenin signaling pathway have been involved in different cancer types, particularly colorectal cancer, osteoporosis, and other human diseases (Clevers & Nusse, 2012; Moon et al. 2002, 2004; Nusse, 2005). The high mortality rate due to cancer can be prevented if this pathway is inhibited.
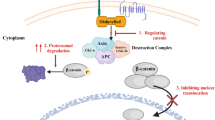
The Wnt signaling pathway can activate both the β-catenin-independent non-canonical pathway and the β-catenin-dependent canonical pathway. When Wnt signaling is absent, the degradation complex, comprising Axin scaffolding protein, tumor suppressor adenomatous polyposis coli (APC), and the (Glycogen synthase kinase 3) GSK 3β, degrades β-catenin, and this prevents the buildup of β-catenin. When Wnt signaling is present, the WNT receptor combines with low-density lipoprotein-receptor related protein 5/6 (LRP5/6) and Frizzled (FZD). This causes the interruption of the degrading complex, which induces the buildup of β-catenin and then moves to the nucleus and combines with the T cell factor/lymphoid enhancer-binding factor (TCF/LEF). Thus, promoting different β-catenin gene activation as shown in Fig 1. Mutations and dysfunction in Wnt/β-catenin signaling component genes such as tumor suppressor gene, β-catenin genes, and adenomatous polyposis coli (APC) are implicated in the constitutive activation of Wnt signaling in more than 90% of colorectal cancer cases (Novellasdemunt et al. 20152015). Hence, there is a need to introduce therapy targeted against the Wnt/β-catenin signaling pathway, which prevents cancerous cell involvement and gets rid of the diseases. Therefore, careful study of the genes involved or unregulated in colorectal cancer will give a clue for the generation of new sustainable therapeutic interventions. Phytochemicals from natural sources possess promising chemoprotective properties, which might serve as reliable and alternative cancer treatments. This study used bioactive compounds from Vitis vinifera to design new inhibitors targeted against β-catenin.
Wnt/β-catenin pathway (Centelles 2012)
Vitis vinifera is a well-known grape species of the family Vitaceae. The different parts of Vitis vinifera contain phytochemical substances, including aromatic acids, flavonoids, phenolic compounds, proanthocyanins, stilbenoids, etc. (Filocamo et al. 2015; Goufo et al. 2020; Radulescu et al. 2020). Research showed that Vitis vinifera’s bioactive constituents have anticancer, antioxidant, antidiabetic, antiviral, and anti-inflammatory potentials (Chaudhary et al. 2021). Vitis vinifera exerts different anticancer properties such as inhibition, apoptosis, cell proliferation, and cell cycle arrest in cancerous cells (Nandakumar et al. 2008).
This study employed a comprehensive computational approach to determine the therapeutic properties of different bioactive compounds from Vitis vinifera. The bioactive compounds were used to impede the Wnt/β-catenin pathway. The study includes molecular docking, ADMET screening, pharmacokinetics, and 3D pharmacophore modeling.
Methods
Ligand library generation and preparation
Secondary metabolites from V. vinifera that have been described were extracted in standard format (SDF) from the online database, Pubchem (Kim et al. 2016). Using the ligPrep tool (Release Schrödinger, 2017), the mined structures were converted into a three-dimensional structure by ionizing at pH (7.2 ± 0.2) and removing salt with Epik (Shelley et al. 2007; Schrodinger, 2021). The OPLS3 force field (Harder et al. 2016) was used for ionization and tautomeric state creation.
Preparation of target
X-ray crystalline structure of β-catenin coupled with an inhibitor (PDB ID: 1JDH) (Asthana et al. 2014) was recovered from Protein Data Bank. The protein was processed with the protein preparation wizard tool of Maestro, Schrodinger Suite. The protein was refined by optimizing the H-bond assignment and minimized using the OPLS3 force field.
Receptor grid generation
The receptor grid depicts the sector where the ligand and protein interact. The Receptor Grid Generation tool was used to create the processed protein grid on the binding site (Glide Grid). Selecting the co-crystallized ligand at the active site of 1JDH revealed the binding location. The ligand on the protein’s crystal structure revealed information about the active site. A cubic grid box including all amino acid residues at the active site was automatically generated with the coordinate X = − 0.360, Y = 14.401, and Z = 47.310, respectively.
Docking
Docking was done on maestro 11.1 (Schrödinger Release 2017) with the Glide tool. Using extra precision (XP) and standard precision (SP) docking algorithm, the crystal structure of β-catenin was utilized to virtually screen the prepared compounds to predict compounds with the lowest docking score. The docking research was carried out with the protein expressed as a rigid body and the ligand’s rotatable bonds set to be free.
To create comparison research, known cancer standards were docked using the same approach.
ADMET/Tox screening
To establish the pharmacokinetic profile, drug-likeness, and toxicity of the hit compounds, the swissADME (http://www.swissadme.ch) and Pro-Tox II online servers (https://tox-new.charite.de/protox II) online servers were used.
MM/GBSA
The molecular mechanics/generalized Born surface area (MM/GBSA) continuum solvent model was employed to discover the docked protein–ligand complex binding free energy.
Rotamer search techniques from prime were used with the VSGB solvent approach and OPLS3 force field to complete this project.
Pharmacophore modeling
The lead compounds’ receptor–ligand complexes were examined. A hypothesis (E-pharmacophore) was generated using the Schrodinger suite’s phase interface to highlight the significant properties that actively contribute to the lead ligands’ characteristic binding to the active sites of the target proteins.
Results
This study features a computational approach to screen phytochemicals of V. vinifera against β-catenin, a therapeutic target protein that takes part in carcinogenesis through the wnt pathway by employing molecular docking, pharmacokinetics screening, MM/GBSA, and pharmacophore modeling approach.
The molecular docking and binding energy study (Figs. 2 and 3) predicted seven compounds as lead compounds that show a better inhibitory potential when compared with the standard compounds, leucovorin Calcium and Xeloda.
Interaction between the lead compounds and the amino acid residues at the active site of the target compared with standards. A—2D interaction between cis-astringin and the amino acid residues at the active site of the target. B—2D interaction between Rutin and the amino acid residues at the active site of the target. C—2D interaction between caftaric acid and the amino acid residues at the active site of the target. D—2D interaction between trans-caftaric acid and the amino acid residues at the active site of the target. E—2D interaction between procyanidin B3 and the amino acid residues at the active site of the target. F—2D interaction between cis-Miyabenol C and the amino acid residues at the active site of the target. G—2D interaction between ampelopsin H and the amino acid residues at the active site of the target. H—2D interaction between leucovorin Calcium and the amino acid residues at the active site of the target. I—2D interaction between Xeloda and the amino acid residues at the active site of the target
A pharmacophore hypothesis which analyzes the characteristic binding of the top-scoring ligands to the target was analyzed and represented in Fig. 4
The lead compounds were subjected to ADME/Tox screening to study their physicochemical properties, pharmacokinetics profile, drug-likeness and toxicity. The results are recorded in Tables 1, 2 and 3.
Discussion
The molecular docking result identified seven compounds from V. vinifera that showed better inhibitory potential against the target enzyme by having better docking scores when compared with the standard compounds such as leucovorin Calcium and Xeloda currently used to manage cancer. A more negative docking score indicates more inhibitory potential. The docking scores, as represented in Fig. 2, show that cis-astringin, rutin, caftaric acid, trans-caftaric acid, procyanidin B3, cis-Miyabenol C, and ampelopsin H show better inhibitory potential by having docking scores of − 7.925, − 6.460, − 6.379, − 6.379, − 6.170, − 5.734 and − 5.548 kcal/mol, respectively, compared to leucovorin Calcium and Xeloda which are standard compounds used to treat cancer with the docking score of − 5.490 and − 3.285 kcal/mol, respectively.
The differences in the lead compounds and standards docking scores are pictorially represented in Fig. 2.
Structural-based drug design is predominantly based on the interaction of protein–ligand, as significant inhibition is primarily determined by the interaction of the ligand with the combination of amino acid residues at the active site of the target enzyme. The interaction which contributes significantly to the inhibition of β-catenin in this study is shown in Fig. 3.
The 2D interaction, as shown in Fig. 3, shows that cis-astringin has hydrogen bonds with GLN 24, GLN 28, TRP 338, ARG 386, ASN 415, and ASN 380, pi-cation with LYS 345, and pi-pi stacking with TRP 383. Rutin forms hydrogen bonds with GLN 24, GLN 28, ARG 386, ASP 459, and ASN 415. Caftaric acid has hydrogen bonds with GLN 375, ASN 380, GLU 28, SER 35, TRP 338, ARG 342, and a salt bridge with LYS 345. Trans-caftaric acid has hydrogen bonds with GLN 375, ASN 380, GLU 28, SER 35, TRP 338, ARG 342, and a salt bridge with LYS 345. Procyanidin B3 shows a hydrogen bond with GLN 24, GLN 28, ASN 387, GLU 33, and ASN 415. It also has pi-pi stacking TRP 383. Cis-Miyabenol C has hydrogen bonds with ASN 380, GLN 379, GLU 33, and SER 31. It also has pi-pi stacking with TRP 383. Ampelopsin H shows hydrogen bonds with GLN 24, GLN 27, ASP 459, SER 32, and ASN 380. It also has pi cation with LYS 345 and pi-pi stacking with TRP 383. Leucovorin Calcium, a standard compound compared with the lead compounds, has hydrogen bonds with SER 35, ARG 342, GLN 28, ARG 386, TRP 338, and ASN 415. It also has a salt bridge with ARG 342, while Xeloda, a standard compound, has hydrogen bonds with ARG 386 and LYS 345.
The MM-GBSA technique is a significantly more accurate way of estimating protein–ligand complexes’ binding free energies (dG) (Bandyopadhyay et al. 2021). It is one of the promising approaches used to increase virtual screening results. A negative dG value suggests that the complexes generated were stable in the target’s binding pocket (Bathula et al. 2021). All lead compounds show a negative dG value, as shown in Fig. 2. The binding free energy of the docked complexes was − 54.285, − 42.282, − 31.078, − 31.078, − 43.693, − 42.348, and − 53.368 kcal/mol for cis-astringin, rutin, caftaric acid, trans-caftaric acid, procyanidin B3, cis-Miyabenol C, and ampelopsin H, respectively. This implies that all the hit compounds are more stable in the target’s binding pocket than the standard compounds leucovorin Calcium and Xeloda, which have − 28.175, and − 24.100 kcal/mol as their free binding energy.
A pharmacophore hypothesis founded on the complex formed was developed to analyze the characteristic binding of the top-scoring ligands to the target. PHASE graphical user interface in Schrodinger’s suite puts forward essential information on the molecular orientation of vital functional groups predominantly involved in the characteristic binding of high-affinity ligands to the protein target (Dixon et al. 2006).
The generated E-pharmacophore hypothesis (Fig. 4) contains H-bond donor, H-bond acceptor interactions, and aromatic rings. This hypothesis can be used to form the basic structural foundation that will be general to all potential inhibitors of the protein target and can also be employed to create a basic skeleton of compounds with a specific angular distance that will bond firmly to the ligand-binding site of the targets.
The absorption, distribution, metabolism, and excretion (ADME) analysis has proven to be an essential component of the drug discovery process. It is a highly economical method for in silico prediction of drug action within a biological system (Kenakin et al. 2016). In this study, we use the SwissADME and Protox-II servers to predict the drug-like and toxicity profile of the selected lead and standard compounds.
The physicochemical properties are established on the Lipinski rule, which considers the five (5) main criteria to determine if a drug is orally active (Lipinski et al. 1997). The results are shown in Table 1. Cis-astringin showed very good physicochemical properties, low molecular weight, and a favorable bioavailability score and solubility. This makes it the top drug-like compound among the test compounds based on physicochemical profiling. While Calcium Xeloda showed a good physicochemical profile with no violation of the Lipinski’s rule, leucovorin has a very poor physicochemical profile, and the molecular weight exceeds the Lipinski’s standard (500 g/mol), the bioavailability score is low, and compared to Cis-astringin, it is a poor compound.
The pharmacokinetic profile of the compounds is shown in Table 2. Ampelopsin H, rutin, and the standard compounds were P-glycoprotein substrates. P-glycoprotein is one of the ATP binding cassette (ABC) proteins involved in discharging molecules from the cell and preventing compounds from bioaccumulating and eliciting their response. (Lin et al. 2003) The low gastrointestinal absorption rate of the test compounds and leucovorin indicates their poor lipophilic nature. However, Calcium Xeloda showed a high rate of gastrointestinal absorption. The test and standard compounds do not permeate the blood–brain barrier. Cytochrome P450 (CYP) is a family of enzymes that catalyze the phase 1 metabolism of xenobiotics at large. Any compound that inhibits selected isoforms would induce a drug–drug interaction. (Esteves et al. 2021). Except for procyanidin B3, which was predicted to inhibit the isoform CYP34A, all of the other test compounds and the standard compounds were expected to be non-inhibitors of these enzymes and these cannot induce a drug–drug interaction.
The Pro-tox II online server estimated the toxicity profile of compounds, and it is shown in Table 3. The least toxic compound is rutin. It has the highest predicted LD50 value of 5000 mg/kg and ranks 5 in the toxicity class. (It ranges from 1 to 6, i.e., most toxic to least toxic.) It is predicted to have no carcinogenic or hepatotoxic effect. Leucovorin has an expected LD50 value of 135 mg/kg and ranks 3 in toxicity, making it the most toxic compound. Although Calcium Xeloda is not predicted to be as toxic as leucovorin, it is a hepatotoxic compound.
Conclusions
This work used in silico analysis to predict seven compounds from V. vinifera phytoconstituents: cis-astringin, rutin, caftaric acid, trans-caftaric acid, procyanidin B3, cis-Miyabenol C, and ampelopsin H as lead compounds that can bind to β-catenin more robustly than the conventional compounds. When these lead compounds are subjected to ADME/Tox screening, it is discovered that they are somewhat drug-like, with cis-astringin being the most drug-like compound compared to the co-crystallized compound.
This shows that these compounds could be better drug-like molecules, and V. vinifera could be a good plant source for a drug-like compound that could cure tumors by suppressing the Wnt//β-catenin pathway’s target enzyme, β-catenin.
Lead compound optimization and additional in vitro and in vitro analyses are recommended to validate this experiment.
Availability of data and materials
All data and materials are available on request.
Abbreviations
- ADMETox:
-
Absorption, distribution, metabolism, excretion, and toxicity
- APC:
-
Adenomatous polyposis coli
- MMGBSA:
-
The molecular mechanics/generalized Born surface area
- TCF/ LEF:
-
T cell factor/Lymphoid enhancer-binding factor
- LRP5/6:
-
Low-density lipoprotein-related protein 5/6
- FZD:
-
Frizzled
- CYP:
-
Cytochrome p450
- ABC:
-
ATP binding cassette
- GSK3:
-
Glycogen synthase kinase 3
- FDA:
-
Food and Drug Administration
- XP:
-
Extra precision
- SP:
-
Standard precision
References
Asthana S, Agarwal T, Banerjee I, Ray S (2014) In silico screening to elucidate the therapeutic potentials of asparagamine A. Homo 3:14
Bandyopadhyay S, Abiodun O, Ogboo B, Kola-Mustapha A, Attah E, Edemhanria L, Kumari A, Jaganathan R, Adelakun N (2021) Polypharmacology of some medicinal plant metabolites against SARS-CoV-2 and host targets: molecular dynamics evaluation of NSP9 RNA binding protein. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1959401
Bathula R, Muddagoni N, Lanka G, Dasari M, Potlapally S (2021) Glide docking, autodock, binding free energy and drug-likeness studies for prediction of potential inhibitors of cyclin-dependent kinase 14 protein in wnt signaling pathway. Biointerface Res Appl Chem 12(2):2473–2488
Centelles JJ (2012) General Aspects of Colorectal Cancer. ISRN Oncology 2012. https://doi.org/10.5402/2012/139268
Chaudhary K, Parihar S, Sharma D (2021) A critical review on nanoscience advancement: in treatment of viral infection. J Drug Deliv Ther 11(6):225–237
Choi B, Cave C, Na C, Sockanathan S (2020) GDE2-dependent activation of canonical wnt signaling in neurons regulates oligodendrocyte maturation. Cell Rep 31(5):107540
Clevers H, Nusse R (2012) Wnt/β-catenin signaling and disease. Cell 149(6):1192–1205
Dixon S, Smondyrev A, Rao S (2006) PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 67(5):370–372
Esteves F, Rueff J, Kranendonk M (2021) The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family. J Xenobiot 11(3):94–114
Filocamo A, Bisignano C, Mandalari G, Navarra M (2015) In vitro antimicrobial activity and effect on biofilm production of a white grape juice (Vitis vinifera) extract. Evid Based Complement Altern Med 2015:1–5
Goufo P, Singh R, Cortez I (2020) A reference list of phenolic compounds (including stilbenes) in grapevine (Vitis vinifera L.) roots, woods, canes, stems, and leaves. Antioxidants 9(5):398
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang J, Wang L, Lupyan D, Dahlgren M, Knight J, Kaus J, Cerutti D, Krilov G, Jorgensen W, Abel R, Friesner R (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296
Kenakin T (2016) Pharmacology in drug discovery and development: understanding drug response. Academic Press, pp 275–299
Kim S, Thiessen P, Bolton E, Chen J, Fu G, Gindulyte A, Wang J (2016) BS The PubChem project. Nucleic Acids Res 44(D1):D1202–D1213
Lin J, Yamazaki M (2003) Role of P-glycoprotein in pharmacokinetics. Clin Pharmacokinet 42(1):59–98
Lipinski C, Lombardo F, Dominy B, Feeney P (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Moon R, Bowerman B, Boutros M, Perrimon N (2002) The promise and perils of Wnt signaling through β-catenin. Science 296(5573):1644–1646
Moon R, Kohn A, Ferrari G, Kaykas A (2004) WNT and β-catenin signalling: diseases and therapies. Nat Rev Genet 5(9):691–701
Nandakumar V, Singh T, Katiyar S (2008) Multi-targeted prevention and therapy of cancer by proanthocyanidins. Cancer Lett 269(2):378–387
Novellasdemunt L, Antas P, Li V (2015) Targeting Wnt signaling in colorectal cancer. A review in the theme: cell signaling: proteins, pathways, and mechanisms. Am J Physiol-Cell Physiol 309(8):C511–C521
Nusse R (2005) Wnt signaling in disease and in development. Cell Res 15(1):28–32
Radulescu C, Buruleanu L, Nicolescu C, Olteanu R, Bumbac M, Holban G, Simal-Gandara J (2020) Phytochemical profiles, antioxidant and antibacterial activities of grape (Vitis vinifera L.) seeds and skin from organic and conventional vineyards. Plants 9(11):1470
Salik B, Yi H, Hassan N, Santiappillai N, Vick B, Connerty P, Duly A, Trahair T, Woo A, Beck D (2020) Targeting RSPO3-LGR4 signaling for leukemia stem cell eradication in acute myeloid leukemia. Cancer Cell 38(2):263-278. e266
Schrödinger Release (2021)-1 Epik, Schrödinger, LLC, New York, NY, 2021; https://www.schrodinger.com/citations
Schrödinger L (2017) Schrödinger, LLC; New Yorkn2017. Schrödinger Suite, 2, 2017-1
Shelley J, Cholleti A, Frye L, Greenwood J, Timlin M, Uchimaya M (2007) Epik: a software program for pKaprediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21(681–691):24
Soleas J, D’Arcangelo E, Huang L, Karoubi G, Nostro M, McGuigan A, Waddell T (2020) Assembly of lung progenitors into developmentally-inspired geometry drives differentiation via cellular tension. Biomaterials 254:120128
Acknowledgments
This research has no acknowledgment.
Funding
No external funding was received for this research.
Author information
Authors and Affiliations
Contributions
AOA was involved in conceptualization and manuscript writing. AOA contributed to result analysis and manuscript writing. OO was involved in manuscript editing and result analysis. PAA and SOO contributed to experimentations and manuscript editing. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adebesin, A.O., Ayodele, A.O., Omotoso, O. et al. Computational evaluation of bioactive compounds from Vitis vinifera as a novel β-catenin inhibitor for cancer treatment. Bull Natl Res Cent 46, 183 (2022). https://doi.org/10.1186/s42269-022-00872-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-022-00872-3