Abstract
Backgrond
In silico studies are essential techniques in the modern medicinal chemistry. QSAR modeling and molecular docking are important techniques in both drug discovery and development and have been successfully deployed in the field of medicinal chemistry for the discovery, design, and development of many drug candidates. These techniques were used in this work to come up with a model that relate 2,5-disubstituted furans with their antiplasmodium activities for the development of more active antimalarial drugs.
Results
Predictive and robust QSAR model was generated using Genetic Function Algorithm. The model was statistically validated to have internal and external squared correlation coefficient, R2 of 0.982 and 0.735 respectively; predictive squared correlation coefficient, R2pred of 0.599; adjusted squared correlation coefficient, Radj of 0.974; and leave-one-out cross-validation coefficient, Q2cv of 0.966. It was found out that the antiplasmodium activities of 2,5-disubstituted furans relied on the parameters: GATS5c, minsCl RDF130m, RDF75p, and RDF115s descriptors. All the descriptors except minsCl influenced the antiplasmodium activities of the compounds negatively. That is, their increase decreases the activities of the furans and vice versa. The docking study revealed that most of the furans bind more tightly to Plasmodium falciparum lactate dehydrogenase (pfLDH) than chloroquine, but the enzyme may not be their major target.
Conclusion
Insight into the relationship between 2,5-disubstituted furans and their antiplasmodium activities has been revealed from the results of this work. Therefore, this could serve as a model for designing novel 2,5-disubstituted furans as potential antimalarial drugs with better activities.
Similar content being viewed by others
Background
World Health Organization reported 228 million cases and 405,000 deaths worldwide from malaria disease in 2018, where children (< 5 years) accounted for 272,000 deaths. This made it one of the most lives threatening disease prevalent in Africa, where 93% and 94% of the 2018 global cases and deaths respectively came from the region (World Health Organization, 2019). Nigeria is the country with the highest malaria burden responsible for 25% and 24% of the 2018 global cases and deaths, respectively (World Health Organization, 2019). Out of the five species of Plasmodium parasite known to cause malaria, Plasmodium falciparum is the most deadly (Cohen et al., 2012) and commonest in the WHO African region responsible for 99.7% of estimated malaria cases of the region in 2018 (World Health Organization, 2019). Therefore, controlling the disease is important to global health.
Malaria is prevented by vector control through the use of treated mosquito nets and insecticides (World Health Organization, 2019). And the disease is treated by chemotherapies using quinoline derivatives (Salas et al., 2013), artemisinin and its derivatives (Jaen et al., 1990), antifolate drugs (Newton & White, 1999), and some antibiotics (Rathore et al., 2005). But, treatment and elimination of malaria remains a global challenge due to the parasite resistance to existing drugs, mosquito resistance to insecticides, and lack of successful malaria vaccine (Singh et al., 2015). WHO recommends the use of artemisinin-based combination therapies (ACTs) as first-line treatment for P. falciparum (Martinelli et al., 2008). However, resistance to ACTs is increasing in South East Asia Region, the Greater Mekong Subregion (GMS), and Western Pacific Region (World Health Organization, 2019). This could spread to other regions of the world, therefore, the need for highly potent antimalaria with low propensity to resistance.
Krake and coworkers studied 2,5-disubstituted furans as novel inhibitors of P. falciparum (Krake et al., 2017). Their study revealed that the furans have good in vitro activities (in EC50) against P. falciparum NF54 strain, low propensity to generate resistance and long half-life which is a good characteristic of antimalaria in accordance with Medicines for Malaria Venture (MMV) objective (Krake et al., 2017). But the analogues had tight Structure Activity Relationship (SAR) limitations, and their activities need improvement. In silico studies are efficient modeling techniques used to screen chemical databases so as to find novel drug leads (Tropsha, 2010). Herein, we carried out quantitative structure activity relationships (QSAR) on the 2,5-disubstituted furans analogues so as to come up with an empirical relationship between the chemical structures of the furans and their antiplasmodium activities, which could be used to design novel 2,5-disubstituted furans with better activities against P. falciparum. We also performed molecular docking study on the furans with Plasmodium falciparum lactate dehydrogenase (PfLDH) as the potential target so as to investigate the mode of interactions of the compounds with the target and have insight into ways of improving their antiplasmodium activities.
Materials and method
Data collection
2,5-Disubstituted furans with their in vitro activities against Plasmodium falciparum NF54 strain were gotten from the paper of Krake and coworkers (Krake et al., 2017) for used in this research. The antiplasmodial activities of the furans obtained as EC50 (nM) were converted to pEC50 (pEC50 = − logEC50) for the purpose of this research. The structures of the furans molecules and their activities are presented in Table 1.
Geometric optimization
The molecules presented in Table 1 were drawn with the aid of Chemdraw (Li et al., 2004), and optimized using B3LYP functional and 6-31G basis set with the aid of the Spartan 14 software Version 1.1.4 (Becke, 1993).
Descriptors computation, normalization and pretreatment
The optimized molecules were imported to the PaDEL-Descriptor software in an SD file format, where 1875 molecular descriptors of the furans molecules were calculated (Yap, 2011). The descriptors were normalized based on Eq. (1) in order to give each variable equal opportunity to influence the construction of a high-quality model (Singh, 2013).
In the equation, X is the normalized descriptors, Xi is the descriptor’s value for each molecule, and Xmin and Xmax are the minimum and maximum values for each descriptor. Redundancy in the normalized data was eliminated using the Data Pretreatment software obtained from Drug Theoretical and Cheminformatics Laboratory (DTC Lab).
Data division
Using the Data Division software of DTC Lab, Kennard Stone method was employed in dividing the pretreated data into training set (70%) and test set (30%) (Kenard & Stone, 1969). The training set was used for model generation and internal validation while the test set was used for external validation.
Model generation
Genetic Function Approximation (GFA) incorporated in the Material Studio software was employed to select descriptors and carry out regression analysis on the training set so as to generate the model using the descriptors and the activities of the furans in pEC50 as the independent and the dependent variable, respectively.
Validation of the model generated
The model generated was assessed using Friedman formula (Friedman, 1991) defined as follows:
LOF is the Friedman’s lack fit which is a measure of fitness of a model; SEE stands for standard error of estimation; p, d, c, and M are total number of descriptors in the model, user-defined smoothing parameter, number of terms in the model, and the number of compound in the training set, respectively.
SEE, the standard deviation of the model whose value has to be low for a good model is defined as follows:
Square correlation coefficient, R2, of a built model is another parameter considered and the closer it is to 1.0, the better the model built. This is expressed as follows:
Yprd, Yexp, and Ymtrn are the predicted, experimental, and mean experimental activities in the training set, respectively.
The value of R2 is directly proportional to the number of descriptors; hence, the stability of the model is not reliable on it. Thus, to have a reliable and stable model, R2 is adjusted according to the expression as follows:
where p is the number of descriptors in the model, and n is the number of compounds used in training set.
Cross-validation coefficient, Q2cv, is expressed as follows:
where Yprd, Yexp, and Ymtrn are the predicted, experimental, and average experimental activity in the training set, respectively.
Square correlation coefficient, R2pred, of the generated model using test set was calculated for external validation. The closer the value is to 1.0, the better the model generated (Tropsha et al., 2003). It is expressed as follows:
Yprd and Yexp are respectively the predicted and experimental activities of the test set, and Ymtrn is the mean experimental activity of the training set.
Descriptors contribution
Degree of contribution of each descriptor in affecting the antiplasmodium activities of the furans was accessed by Mean Effect (MFj) value expressed as follows:
bj is the coefficient of descriptor j in the model, dij is the value of the descriptor j in the descriptor matrix for each molecule in the training set, and m and n are the number of descriptors in the model and the number of training set molecules (Habibi-Yangjeh & Danandeh-Jenagharad, 2009). Variance inflation factor, VIF, and correlation analysis were performed on the descriptors in other to check for multi-co-linearity problem.
Y-randomization test
Random multi-linear regression models were generated (using training set) in Y-randomization test whose R2 and Q2 values have to be low for the QSAR model to be robust (Tropsha et al., 2003). Coefficient of determination, cR2p, whose value has to be greater than 0.5 for passing this test is also calculated in the Y-randomization test and is expressed as follows:
R is the correlation coefficient for Y-randomization, and R2r is the average “R” of the random models.
Applicability domain of the generated model
The applicability domain of the model was determined using leverage (hi) (Veerasamy et al., 2011) and is expressed for a compound, i, as follows:
Xi is the training compounds matrix of i. X is the n × k descriptor matrix of the training set compound, and XT is the transpose matrix of X used to generate the model. The warning leverage, h*, is the maximum value for X and is expressed as follows:
n is the number of training compounds, and p is the number of descriptors in the model.
Molecular docking
Optimized structures of chloroquine and nine furans molecules having the best antiplasmodium activities were saved as PDB file from the Spartan software and exported to the pyrex software were they were converted to PDBQT file. Crystal structure of the enzyme, Plasmodium falciparum lactate dehydrogenase (pfLDH) was obtained from protein data bank (PDB ID: 1CET) (Read et al., 1999), prepared by removing water molecules, co-crystallized ligand and hetero-atoms using the Discovery Studio software, then saved as PDB file and exported to the pyrex software were it was converted to PDBQT file. The prepared furan molecules (the ligands) and the prepared enzyme (the receptor) were docked using Autodock Vina in the Pyrx software after assessing the reliability of the protocol by re-docking of the enzyme with chloroquine (Trott & Olson, 2010).
Results and discussion
Genetic Function Algorithm (GFA) was used to generate QSAR model that relate the chemical structure of 2,5-disubstituted furans with their biological activities as active antimalarials. The model is presented as follows:
A predictive, robust, and reliable QSAR model that passed all the validation criteria was generated. The validation parameters of the model were given in Table 2. The model contained autocorrelation (GATS5c), atom type electro-topological (minsCl), and radial distribution function (RDF130m, RDF75p, and RDF115s) descriptors. GATS5c is a 2D Geary autocorrelation of lag 5 weighted by gasteiger charge. Here, gasteiger charge is the physico-chemical property calculated for every atom in the molecule and is the Geary coefficients. Increase in GATS5c decreases the pEC50 of the furans and vice versa. Minimum atom-type E-State: -Cl, minsCl is a 2D descriptor which showed the important of the present of chlorine atoms in the furans molecules. This descriptor incorporated the steric and electronic effects of the atoms surrounding any chlorine atom in the molecules (Kier & Hall, 1999), and it had positive contribution to the model; hence, increase in the descriptor increases the antiplasmodium activities of the furans. The third class of descriptors in the model was a radial distribution function descriptor denoted as RDFЅω where ω is any measurable molecular property, and S is the radius from the geometrical center of the molecule at which ω is measured for any atom or group of atoms in the molecule. S and ω for the descriptors RDF130m, RDF75p, and RDF115s were atomic mass, atomic polarizability, and electronic inductive effect (I-state) and 13 Å, 7.5 Å, and 11.5 Å, respectively. These descriptors contributed negatively to the antiplasmodium activities of the furans that is increase in them decreases the activities of the furans. The power of model in predicting the antiplasmodium activities of the furans is indicated by the low residual values (Table 3).
Table 4 presents the Pearson’s correlation matrix, variance inflation factor and Mean Effect of the five descriptors in the model. The highest coefficient in the correlation matrix was 0.465085 between the descriptors RDF75p and minsCl which showed no significant inter-correlation among the descriptors in the model, and the variance inflation factor values for all the descriptors were less than 2. These mean that the descriptors in the model were good and void of multi-co-linearity (Beheshti et al., 2016).The Mean Effect revealed the extent to which each descriptor contributed to the antiplasmodium activities of the compounds. Therefore, the most important descriptor in the model was GATS5c, and the least was minsCl. The average R, R2, Q2, and the cRp2 from the Y-randomization test result presented in Table 5 confirmed the reliability, robustness, and stability of the built QSAR model (Tropsha et al., 2003; Roy et al., 2016). Hence, the model was not gotten by chance.
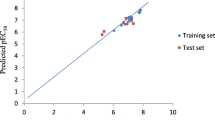
The plot of predicted against experimental activities of the compounds as presented in Fig. 1, which indicated the predictive power of the generated model as shown by the linearity of the plot. Therefore, the model is good in predicting the antiplasmodium activities of 2,5-disubstituted furans scaffold. The generated model is void of systematic error as shown by the distribution of standardized residual on both sides of zero (Fig. 2) (Jalali-Heravi & Kyani, 2004). The plot of standardized residuals versus leverages (Williams plot) is given in Fig. 3. It showed that all the training and test set compounds are within the applicability domain as their leverage values was less than the warning leverage (h∗ = 1.125) and their standardized residuals within ± 2.5. Therefore, there is no outlier or influential compound; hence, any of the compound can be used in designing new potent 2,5-disubstituted furans molecules.
The docking result shows that out of the nine docked furans, five had greater binding affinity, and almost all had greater interactions with the enzyme than chloroquine (Table 6). The compounds bind to the receptor via different amino acid; this suggests that they have different mechanisms of action. Compound 22 has the highest binding energy of − 6.9 kcal/mol among the docked compounds. This compound formed two conventional hydrogen bonds between the oxygen atom of the furan moiety and the HN fragment of MET30 and ILE31 amino acids of the enzyme, one conventional hydrogen bond between hydrogen atom from alkyl substituent of the furan and oxygen atom of THR97 residue, and one carbon hydrogen bond between carbon atom from alkyl substituent of the furan and oxygen atom of GLY99 residue (Fig. 4). The compound also formed hydrophobic interactions with the receptor: two of pi-sigma type with ILE31 and ILE31 residues, two of alkyl-alkyl type with ALA236 and PRO246 residues, and one of pi-alkyl type with PRO246 residue. Figure 5 shows the hydrogen bonds and hydrophobic bonds surfaces of the interactions. Although the binding affinities of the docked compounds did not correlate well with their antiplasmodium activities suggesting that pfLDH may not be their target enzyme or rather their main target enzyme, the results could be utilized in developing highly potent furan molecules capable of inhibiting pfLDH.
Conclusion
QSAR model that linked the chemical structures of 2,5-disubstituted furans and their antiplasmodium activities was generated using Genetic Function Algorithm in the Material Studio software. The model was statistically validated to be reliable and robust with internal and external square correlation coefficient of 0.982 and 0.735, respectively. The model revealed that GATS5c, minsCl RDF130m, RDF75p, and RDF115s descriptors connected to the antiplasmodium activities of the furans where increase in all, but minsCl descriptors decrease the activities and vice versa. Molecular docking study conducted revealed that the furans interacted well with pfLDH enzyme, but the enzyme may not be their major target as the binding energy of the docked compound did not correlate well with their antiplasmodium activities. The QSAR and the molecular docking studies could be married together to design novel furans molecule with high potency to inhibit Plasmodium falciparum.
Availability of data and materials
All the data generated during the current study are included in this published article, and the datasets used during the study are available from the article:
Krake SH, Martinez PDG, McLaren J, Ryan E, Chen G, White K, Charman SA, et al. Novel inhibitors of Plasmodium falciparum based on 2,5-disubstituted furans. Eur J Med Chem. 2017; doi: 10.1016/j.ejmech.2016.12.024.
Abbreviations
- WHO:
-
World Health Organization
- MMV:
-
Medicines for Malaria Venture
- QSAR:
-
Quantitative structure activity relationships
- GFA:
-
Genetic Function Algorithm
- pfLDH:
-
Plasmodium falciparum lactate dehydrogenase
- EC50 :
-
Half maximal effective concentration
- ME:
-
Mean Effect
- VIF:
-
Variance inflation factor
References
Becke AD (1993) Becke’s three parameter hybrid method using the LYP correlation functional. J Chem Phys. 98:5648–5652
Beheshti A, Pourbasheer E, Nekoei M, Vahdani S (2016) QSAR modeling of antimalarial activity of urea derivatives using genetic algorithm-multiple linear regressions. J Saudi Chem Soc. 20:282–290
Cohen JM, Smith DL, Cotter C, Ward A, Yamey G, Sabot OJ, Moonen B (2012) Malaria resurgence: a systematic review and assessment of its causes. Malar J. 11:122
Damme SV, Bultinck P (2007) A new computer program for QSAR-analysis: ARTEQSAR. J Comput Chem. 28:1924–1928
Eriksson L, Jaworska J, Worth AP, Cronin MT, McDowell RM, Gramatica P (2003) Methods for reliability and uncertainty assessment and for applicability evaluations of classification and regression-based QSARs. Environ Health Perspect. 111:1361
Friedman JH (1991) Multivariate adaptive regression splines. Ann Stat.:1–67
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model. 20:269–276
Habibi-Yangjeh A, Danandeh-Jenagharad M (2009) Application of a genetic algorithm and an artificial neural network for global prediction of the toxicity of phenols to Tetrahymena pyriformis. Monatsh Chem-Chem Mon. 140:1279–1288
Jaen JC, Wise LD, Caprathe BW, Tecle H, Bergmeier S, Humblet CC, Heffner TG, Meltzer LT, Pugsley TA (1990) 4-(1, 2, 5, 6-Tetrahydro-l-alkyl-3-pyridinyl)-2-thiazolamines: a novel class of compounds with central dopamine agonist properties. J Med Chem. 33:311–317. https://doi.org/10.1021/jm00163a051
Jalali-Heravi M, Kyani A (2004) Use of computer-assisted methods for the modeling of the retention time of a variety of volatile organic compounds: a PCA-MLR-ANN approach. J Chem Inf Comput Sci. 44:1328–1335
Kenard RW, Stone LA (1969) Computer aided design of experiments. Technometrics. 11(1):137–148. https://doi.org/10.1080/00401706.1969.10490666
Kier LB, Hall LH (1999) Molecular structure descriptors: the electrotopological state. Academic Press, New York
Krake SH, Martinez PDG, McLaren J, Ryan E, Chen G, White K, Charman SA et al (2017) Novel inhibitors of Plasmodium falciparum based on 2,5-disubstituted furans. Eur J Med Chem. https://doi.org/10.1016/j.ejmech.2016.12.024
Li Z, Wan H, Shi Y, Ouyang P (2004) Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J Chem Inf Comput Sci. 44:1886–1890
Martinelli A, Moreira R, Cravo PVL (2008) Malaria combination therapies: advantages and shortcomings. Mini Rev. Med. Chem. 8:201–212
Newton P, White N (1999) Malaria: new developments in treatment and prevention. Annu Rev Med 50:179–192. https://doi.org/10.1146/annurev.med.50.1.179
Rathore D, McCutchan TF, Sullivan M, Kumar S (2005) Antimalarial drugs: current status and new developments. Expert Opin Investig Drugs. 14:871–883. https://doi.org/10.1517/13543784.14.7.871
Read JA, Wilkinson KW, Tranter R, Sessions RB, Brady RL (1999) Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase. J Biol chem. 274(15):10213–10218
Roy K, Das RN, Ambure P, Aher RB (2016) Be aware of error measures. Further studies on validation of predictive QSAR models. Chemom Intell Lab Syst. 152:18–33
Salas PF, Herrmann C, Orvig C (2013) Metalloantimalarials. Chem Rev. 113:3450–3492. https://doi.org/10.1021/cr3001252
Singh AK, Rajendran V, Pant A, Ghosh PC, Singh N, Latha N, Garg S et al (2015) Design, synthesis and biological evaluation of functionalized phthalimides: a new class of antimalarials and inhibitors of falcipain-2, a major hemoglobinase of malaria parasite. Bioorg Med Chem. 23:1817–1827. https://doi.org/10.1016/j.bmc.2015.02.029
Singh P (2013) Quantitative structure-activity relationship study of substituted-[1, 2, 4] oxadiazoles as S1P1 agonists. J Curr Chem Pharm Sci. 3
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inform. 29:476–488
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. Mol Inform. 22:69–77
Trott O, Olson AJ (2010) Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput chem. 22:455–461
Veerasamy R, Rajak H, Jain A, Sivadasan S, Varghese CP, Agrawal RK (2011) Validation of QSAR models-strategies and importance. Int J Drug Des Discov. 3:511–519
World Health Organization, World Malaria Report, 2019. https://www.who.int/news-room/feature-stores/detail/world-malaria-report-2019. Accessed 26 Jan 2020.
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 32:1466–1474
Acknowledgements
The authors wish to acknowledge the moral support of Kaduna polytechnic Nigeria.
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
AWM and GAS conceived and designed the study. AWM drew and optimized the structures. AWM calculated the descriptors, generated the model, performed the statistical analysis, conducted the molecular docking, and wrote the paper. GAS AU supervised the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahmud, A.W., Shallangwa, G.A. & Uzairu, A. In silico studies of 2,5-disubstituted furans as active antimalarial drug candidates. Bull Natl Res Cent 44, 77 (2020). https://doi.org/10.1186/s42269-020-00333-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-020-00333-9