Abstract
Background
Due to rapid development of microbial resistance against chemotherapeutic agents (mostly antibiotics), it has become essential currently to screen effective, safe, cheap, and available therapeutics from various medicinal plants—like herbs—for their potential antimicrobial effect.
Aim
To estimate the antibacterial activity of aqueous, ethanol, and methanol extracts of each of Moringa oleifera L. leaves and Matricaria recutita L. flowers against antibiotic-resistant and sensitive bacterial strains isolated from patients having wound infections.
Results
In the present study, a total of one hundred clinical samples were obtained from different cases of infected wounds. Forty isolates (40%) of pure bacterial cultures were detected. Pseudomonas aeruginosa was found to be the predominant agent isolated from the wound infections (32.5%) followed by Staphylococcus spp. (25%), E. coli (20%), Klebsiella spp. (20%), and Proteus mirabilis (2.5%). Sensitivity of the bacterial isolates was tested against antibiotic discs: piperacillin, ampicillin, oxacillin, pinicillin, gentamicin, tobramycin, amikacin, streptomycin, ceftriaxone , ceftazidime, cefoxitin, cefoperazone, cefuroxime, cefepime, cefotaxime, ciprofloxacin, oflaxacin, levofloxacin, lomefloxacin, moxifloxacin, tetracycline, aztreonam, azithromycin, erythromycin, imipenem, piperacillin-tazobactum, ampicillin-sulbactam, linezolid, teicoplanin, trimethoprim-sulphamethoxazole , chloramphenicol, and clindamycin. Out of the 40 bacterial strains studied, 20 isolates were multidrug-resistant (MDR), 7 extensively drug-resistant (XDR) and 3 were pan drug-resistant (PDR). The in vitro susceptibility test showed that the water, ethanol (95%), and methanol (80%) extracts of Moringa oleifera L.(leaves) and Matricaria recutita L.(flowers) produced an inhibitory effect against 12 resistant MDR, XDR, and PDR test isolates, with minimum inhibitory concentration (MIC) ranging from 7.8–62.5 mg/ml.
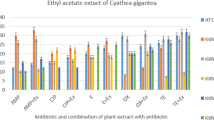
Water and methanol extracts of both plants represented good activity against most of the sensitive and resistant isolates whereas ethanol extract of both plants showed a lesser activity against nearly all of the isolates
Conclusion
This study had the potential value to develop antibacterial agents against resistant (MDR, XDR, and PDR) and susceptible bacteria supporting the significant use of plant extracts in treating wound infections related to bacteria and these active extracts will provide useful information for discovering new compounds with better activity and more effective against resistant (MDR, XDR, and PDR) and susceptible bacteria responsible for wound infections than currently available antibiotic agents.
Similar content being viewed by others
Background
Surgical site infection is the most common hospital-acquired infections in developing countries. Other wound infections include burn wound infection, diabetic foot ulcer infection, bite wound infection, acute soft tissue infection, and pressure ulcer infection (Bhalchandra et al., 2018). Gram-positive cocci such as Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus spp. and Gram-negative bacilli such as Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Proteus species are the most common pathogenic bacteria isolated from wounds (Pallavali et al.,2017).
Antibiotic resistance among bacterial strains is a serious situation. It may be so rapid that the effectiveness of common antibiotics may be lost within a span of 5 years due to genetic changes (Chandra et al., 2017). Pseudomonas aeruginosa is responsible for burns and wound infections; it is also an important cause of wound infections in diabetic individuals and infected wounds following surgeries (Bassetti et al.,2018). Staphylococcus aureus is the most common cause of hospital-acquired wound infections. Klebsiella spp. and E. coli as predominant bacteria are also associated with burn wounds. Multidrug-resistant (MDR) was defined as such because of their in vitro resistance to more than one antimicrobial agent in three or more antimicrobial categories. Extensively drug-resistant (XDR) was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e., bacterial isolates remain susceptible to only one or two antimicrobial categories). To characterize a bacterium as pan drug-resistant (PDR), it must be tested and found to be resistant to all approved and useful antimicrobials (Magiorakos et al., 2012).
One of the surveys conducted by the World Health Organization (WHO) reports that more than 80% of the world’s population still depends upon the traditional medicines for various diseases. Forced with the growing resistance of MDR microbe strains to antibiotics and other drugs, the search for alternatives is serious (WHO, 2005; Chandra et al., 2017 and Dan et al., 2018).
There are numerous plants and natural products which have antibacterial, antifungal, and antiprotozoal effect that could be used either systemically or locally. Medicinal properties of plants have also been preferred throughout the world, due to their potent pharmacological activities, low toxicity, and economic viability, when compared with synthetic drugs. Medicinal plants are rich in a wide variety of bioactive secondary metabolites such as tannins, terpenpoids, alkaloids, saponins, flavonoids, and phenolic compounds that can produce a definite physiological action on the human body (Shakya, 2016).
Moringa oleifera L. is one of the best known, widely distributed, and grown species of a monogeneric family Moringaceae. The plant has been reported to possess antimicrobial properties and this explains the reason for its wide use in the treatment of human diseases. Significant medicinal properties of the plant include anti-inflammatory, antibacterial, and antifungal effects (Abalaka et al.,2012 and Tirado-Torres et al.,2019). Matricaria recutita L. or Matricaria chamomilla L., commonly known as chamomile, is an annual plant of the composite family Asteraceae. It is widely used and well-documented medicinal plants. It was included in the pharmacopeia of 26 countries. It has been used as a medicinal plant for external wounds, eczema, skin irritations, leg ulcers, diaper rash, inflammation of the skin, bacterial skin diseases, and many others (e.g., Ali and Alattar, 2018).
The present study is an attempt to estimate the antibacterial activity of aqueous, ethanol, and methanol extracts of Moringa oleifera L. leaves and Matricaria recutita L. flowers against antibiotic-resistant and sensitive bacterial strains isolated from patients having wound infections.
Material and methods
Plant collection
Fresh flowers of Matricaria recutita L. (Family: Asteraceae) (German chamomile) and fresh leaves of Moringa oleifera L. (Family:Moringaceae) were collected in June to September 2018 from a farm in El-Fayoum governorate, Egypt. They were identified as Matricaria recutita (German Chamomile) and Moringa oleifera by specialists in the Horticulture Research Institute, Agriculture Research Center in EL-Dokky,Giza, Egypt, and in the Herbarium of Cairo University ,Cairo, Egypt. Fresh plant materials were air-dried for 2 weeks and grinded into fine powdered form, by using a grinder, kept in plastic bags, and subjected later to extraction.
Bacterial samples
One hundred bacterial samples were collected from patients with different wound infections. All of these samples were obtained from the Kasr El Aini Hospital, Cairo, Egypt. Wound surface was cleansed with sterile normal saline, then samples were collected using a sterile cotton swabs; the inner surface of the infected area was swabbed gently; swabs were inserted immediately into a tube containing nutrient broth media then transferred to the Microbiology Laboratory unit at Research Institute of Ophthalmology, Giza, Egypt as soon as possible for further investigations (Manikandan and Amsath, 2013).
Antibiotic discs (Oxoid)
For Gram-negative bacteria, the following antibiotic discs were used: penicillins group: piperacillin (PRL), ampicillin (AMP); the aminoglycoside group: gentamicin (CN), amikacin (AK), streptomycin(S), tobramycin (TOB); cephems including the cephalosporin group: ceftriaxone (CRO), ceftazidime (CAZ), cefoxitin (FOX), cefoperazone (CFP),cefuroxime (CXM), cefepime (FEP), cefotaxime (CTX); the fluoroquinolone group: ciprofloxacin (CIP), oflaxacin (OFX), levofloxacin (LEV), lomefloxacin (LOM), tetracycline (TE); monobactams: aztreonam (ATM); the macrolide group: azithromycin (AZM); the carbapenem group: imipenem (IMP); β-lactamase inhibitor combinations: piperacillin-tazobactam (TZP), ampicillin-sulbactam (SAM); and the lipopeptide group: polymyxin B (PB). The antibiotic discs tested for Gram-positive bacteria were as follows: the aminoglycoside group: gentamicin (CN), amikacin (AK); the fluoroquinolone group: ciprofloxacin (CIP), ofloxacin (OFX), moxifloxacin (MFX), tetracycline (TE); the oxazolidinone group: linezolid (LZD), glycopeptides, teicoplanin (TEC); the macrolide group: erythromycin (E), azithromycin (AZM); folate pathway inhibitor: trimethoprim-sulphamethoxazole (SXT); the phenicol group; chloramphenicol (C), lincosamides ; clindamycin (DA); penicillinase-stable penicillins: oxacillin (OX); and penicillinase-labile penicillins: penicillin(P).
Preparation of plant extracts
Water extract
The extraction procedure was performed according to Rahman et al. (2009). Known weight (100 g) air-dried powder from each plant was extracted with 400 ml distilled water by percolation with occasional shaking for 7 days, then filtered and lyophilized under reduced pressure and 200 mg of the dry extracts was dissolved in 0.4 ml of dimethyl sulfoxide (DMSO) to give 500 mg/ml concentration. The dried extract was kept at – 4 °C for antibacterial activity assay.
Organic solvent extraction
Ethanol and methanol extracts: 100 g of air-dried plant powder was extracted by ethanol (95%) and methanol (80% v/v). The residue ( the mixture ) was transferred into a percolator with overnight maceration; the extract was filtered and evaporated to dryness at 40 °C in a water bath. Finally, the dry extract was weighted and the concentration of each extract was calculated. The obtained extracts were stored at − 20 °C for antibacterial activity assay. (There is no difference between the extraction process of ethanol and methanol.)
Identification of bacterial isolates
Identification of the collected Gram-positive and Gram-negative isolates was carried out according to Bergey’s Manual of Systematic Bacteriology (1989) and Cheesbrough (1984).
Preparation of inoculums
Inoculums were standardized to give a density of 106 colony-forming units (CFU)/ml. A loopful of the test organism was inoculated into 5.0 ml of nutrient broth and incubated at 3 °C for 24 h. 0.2 ml from the 24-h culture of the organism was dispensed into 20 ml sterile nutrient broth and incubated for 3–5 h to standardize the culture to 106 CFU/ml (corresponding to 0.5 McFarland standards). Plates were inoculated within 15 min of standardizing the inoculum, to avoid changes in inoculum density (Abalaka et al., 2012).
Antibiotic sensitivity test
The sensitivities of the isolated bacterial species against different antibiotics were tested based on the disc diffusion (Kirby–Bauer) technique (Bauer et al., 1966) as described by Saif et al. (2017).
Antibacterial assay
It was carried out by agar well diffusion method as described by Das et al. (2013). One hundred microliters (106 CFU /ml) fresh microbial culture was spread on a Muller Hilton agar plate with non-toxic swab. Four wells of 6-mm diameter were punched off into the agar medium with sterile cork-borer (6 mm) and filled with 100 μl (500 mg/ml) of plant extract by using a micropipette in each well under aseptic conditions. DMSO was used as a negative control. The plates were allowed to stand for 1 h to allow for pre-diffusion of the extract into the medium. The plates were incubated aerobically in an upright position at 37 ± 2 °C for 24–48 h. The antibacterial screening was evaluated by measuring the zone of inhibition (mm).
Determination of minimum inhibitory concentration and minimum bactericidal concentration
Minimum inhibitory concentration (MIC) is the lowest concentration of the drug which will inhibit growth as measured by observed turbidity in the test tube (CLSI, 2016). The MIC was determined for the antibacterial most efficient extracts, using the method of Greenwood (1989) as described by Usman et al.(2014) .Six sterile test tubes were arranged in four rows, each extract in one row. Each potential extract was determined by micro-broth dilution technique. One hundred microliters of sterile nutrient broth was pipetted into all the tubes.
Two hundred milligrams of dried extract was added to 0.4 ml of dimethyl sulfoxide DMSO to obtain a concentration of 500 mg/ml. Then, 100 μl was used containing 250 mg. Thereafter, there was a serial dilution of the extract in each tube to obtain concentrations of 125, 62.5, 31.25, 15.62, and 7.81 mg ml−1, respectively. One hundred microliters of 106 CFU/ml of each of the tested bacterium were pipette into each test tube and incubated at 37 °C for 24 h. Two control tubes were used: nutrient broth inoculated with bacteria was used as a positive control and nutrient broth containing the plant extract was used as a negative control.
The lowest concentration that kills the organisms completely, where no bacterial growth is observed (MBC) CLSI (2016), was determined by assaying the test tubes resulting from MIC determinations. A loopful of the content of each test tube was inoculated by streaking on a solidified nutrient agar plate and then incubated at 37 °C for 24 h and observed for bacterial growth (Usman et al., 2014).
Results
From one hundred specimens collected, 40 (40%) yielded pure bacterial culture. The distribution of bacterial isolates from the wounds was as follows: 10 (25%) Gram positive cocci (GPC) (Staphylococcus spp.) and 30 (75%) Gram-negative bacilli (GNB) (Fig 1). The detected organisms were 13 (32.5%) Pseudomonas aeruginosa, 10 (25%) Staphylococcus spp, 8 (20%) E. coli, 8 (20%) Klebsiella spp., and only one (2.5%) isolate Proteus mirabilis (Table 1). P. aeruginosa strains showed high sensitivity to levofloxacin (92.3%), polymyxin B (84.6%), and ciprofloxacin (69.23%), and were mostly resistant to aztreonam and ceftazidime (53.8%) and aminoglycosides (Table 2). In the same table, P. aeruginosa isolates: Ps 5, 7, 9 and Ps 10 were variant (sensitive or resistant) against fluoroquinolones (CIP, OFX, and LEVO). The previously tested isolates were also resistant against cephems (CAZ) and monobactams (ATM).
E. coli were mostly sensitive to imipenem, cefoxitin, ceftazidime, aztreonam, and amikacin (87.5%) followed by gentamycin, ceftriaxone and azithromycin (75%), while 100% resistant to cefuroxime, and (87.5%) to ciprofloxacin, levofloxacin, lomefloxacin, oflaxacin, ampicillin- sulbactam, ampicillin, piperacillin, tetracyclin, and cefotaxime (Table 3).
Table 3 also shows that Klebsiella spp. were very sensitive to imipenem and cefoxitin (75%) followed by tobramycin (62.5%), but (100%) resistant to azithromycin followed by cefuroxime, ampicillin-sulbactam, tetracycline (87.5%) and showed (75%) resistance against cefotaxime, fluoroquinolones, ampicillin, and Piperacillin. Proteus mirabilis showed (100%) sensitivity to cefoxitin, ceftazidime, ceftriaxone, cefotaxime, amikacin, tobramycin, ciprofloxacin, levofloxacin, lomefloxacin, and oflaxacin and aztreonam, on the other hand, 100% resistant against tetracycline, azithromycin, and cefuroxime.
Staphylococcus spp. isolates exhibited 100%, 80%, and 60% resistance against penicillin, oxacillin, and tetracyclin respectively. These strains showed 80% sensitivity to chloramphenicol, trimethoprim-sulphamethoxazole, amikacin, ciprofloxacin, oflaxacin, and linezolid and also exhibited 70–60% sensitivity to moxifloxacin, teicoplanin, azithromycin, and clindamycin (Table 4).
Out of the 40 bacterial strains, 20 (50%) were MDR, 7 (17.5%) were XDR, and 3 (7.5 %) were PDR (Fig. 2). Two P. aeruginosa strains were MDR, and 5 were XDR. Six E. coli strains were MDR and one responded as PDR. Out of the 8 Klebsiella spp., 4 strains were MDR, 2 XDR, and 2 were PDR. Proteus mirabilis strain reacted as an MDR, and finally, 7/10 isolates of the Gram-positive Staph.spp. were MDR (Fig. 3).
Water and methanol extracts represented good activity against most of the sensitive and resistant isolates while the ethanol extract of both plants showed a lesser activity against nearly all of the isolates (Table 5). Table 6 shows the MIC and MBC of 12 resistant isolates for water and methanol extracts of M. oleifera leaves and Metricaria recutita flowers. High MIC value was recorded for Klebsiella spp. with 15.6 to 62.5 mg/ml and S. aureus and E. coli with 7.8 to 62.5 mg/ml. On the other hand, lower MIC values were observed for P. aeurogenosa and Proteus mirabilis with 7.8 to 31.25 mg/ml and 15.6 to 31.25 mg/ml respectively. The extracts showed the same bacteriostatic activity against S. aureus and E. coli in the range of 15.6 mg/mL to 125 mg/ml. The MBC of the extracts presented 31.25 mg/ml to 125 mg/ml against Klebsiella spp., 15.6 to 62.5 mg/ml, and 31.25 to 62.5 mg/ml against P. aeurogenosa and Proteus mirabilis respectively.
Discussion
From one hundred specimens collected, 40 (40%) yielded pure bacterial culture. The distribution of bacterial isolates from the wounds were as follows: 10 (25%) bacterial strains were Gram-positive cocci (GPC) and 30 (75%) were Gram-negative bacilli (GNB)( Fig. 1). These findings are in line with those of previous studies in Asia and other African settings (Osariemen et al.,2013). This might be due to the high antibiotic resistance of Gram-negative bacteria .
Most Gram-positive bacteria are surrounded by a coarse peptidoglycan cell wall. This structure, although mechanically strong, appears to offer little resistance to the diffusion of small molecules such as antibiotics (Nikolaidis et al., 2014). Escherichia coli, in contrast, as Gram-negative bacteria, surround themselves with a second membrane, the outer membrane, which functions as an effective barrier.
The high incidence of P. aeruginosa infection recorded in this work is in agreement with other reports by Aljanaby and Aljanaby (2018). Other studies by Sultana et al. (2015) and Mohammed et al.(2017) reported that S. aureus was the most common bacteria isolated from wound infections. These variations could be attributed to numerous factors including the nature of the wound site, and the type of prophylactic antibiotics used for treatment. The data outlined in (Table 2) indicated that P. aeruginosa strains were highly sensitive to levofloxacin (92.3%) followed by polymyxin B (84.6%) and ciprofloxacin (69.23%), however showed resistance against aztreonam and ceftazidime (53.8%). This was explained by Aldred et al. (2014) that the individual members of fluoroquinolones demonstrate different spectra of activity and pharmacokinetic profiles, fluoroquinolones target two essential bacterial enzymes, DNA gyrase (topoisomerase II) and DNA topoisomerase IV. During this process, the drugs trap a reaction intermediate containing quinolone enzyme and broken DNA, which leads to the blockage of DNA replication, and for some bacteria, death occurs within hours. Furthermore, polymyxins bind to the cell membrane and alter its structure. The result of this process causes an increase in the permeability of the cell envelope consisting of the cell wall and the cytoplasmic membrane, leakage of cell contents, and, subsequently, cell death (Parija, 2012). In the same table, P. aeruginosa isolates: Ps 5, 7, 9 and Ps 10 were variant (sensitive or resistant) against fluoroquinolones (CIP, OFX, and LEVO). Resistance is due to changes in DNA gyrase enzyme and/or the topoisomerase enzyme(s) or by the defective function of porine channels (Hooper and Jacoby,2015). The abovementioned strains showed resistance against aminoglycosides (CN, AK, TOB). Aminoglycoside-inactivating enzymes reduced the uptake of the aminoglycoside into bacteria. The previously tested isolates were also resistant against cephems (CAZ) and monobactams (ATM). This resistance is due to β-lactamase production (extended-spectrum beta lactamases) that hydrolyze these drugs (Palzkill, 2018).
Earlier studies by Saha et al. (2017) and Bhalchandra et al.(2018) reported that P. aeruginosa strains were sensitive to polymyxinb, levofloxacin, and ciprofloxacin and showed high resistance against Aztreonam and ceftazidime. Yakha et al. (2015) and Perimet al. (2015) described high resistance against ceftazidime and polymyxin B.
In the present study, Table 3 presents that E.coli strains showed high sensitivity to imipenem, cefoxitin, ceftazidime, aztreonam, and amikacin reached 87.5% followed by gentamycin, ceftriaxone, and azithromycin (75%) . These strains exhibited 100% resistance against cefuroxime and (87.5%) against ciprofloxacin, levofloxacin, lomefloxacin, oflaxacin, beta-lactamase inhibitor combination (ampicillin-sulbactam), ampicillin, piperacillin, tetracycline, and cefotaxime. Similar observations have been reported by Yakha et al.(2015); Gomatheswari and Jeyamurugan (2017), and Sahaet al.(2017) that imipenem, amikacin, and gentamycin were very effective drugs against E. coli isolates but presented resistance against cefuroxime,ciprofloxacin , ofloxacin ,cefotaxime, and ampicillin.
Table 3 also shows that Klebsiella spp. strains were very sensitive to imipenem and cefoxitin (75%) followed by Tobramycin (62.5%). On the other hand, these strains were (100%) resistant to azithromycin followed by cefuroxime, ampicillin-sulbactam, and tetracycline (87.5%) and showed (75%) resistance against cefotaxime, fluoroquinolones, ampicillin, and piperacillin. These results go with the report by Gomatheswari and Jeyamurugan (2017).
In the same table, Proteus mirabilis showed (100%) sensitivity to cefoxitin, ceftazidime, ceftriaxone, cefotaxime, amikacin, tobramycin, ciprofloxacin, levofloxacin, lomefloxacin, and oflaxacin and aztreonam, on the other hand, 100% resistant against tetracycline, azithromycin and cefuroxime. Kassam et al. (2017) stated that Proteus spp. was sensitive to cefoxitin, ceftazidime, ceftriaxone, amikacin, and ciprofloxacin and resistant against tetracycline.
The results in Table 4 illustrated that Staphylococcus spp. isolates exhibited 100%, 80%, and 60% resistance against penicillin,oxacillin, and tetracyclin respectively. These strains showed 80% sensitivity to chloramphenicol, trimethoprim-sulphamethoxazole, amikacin, ciprofloxacin, oflaxacin, and linezolid. These species also exhibited 70–60% sensitivity to moxifloxacin, teicoplanin, azithromycin, and clindamycin. Our study agrees with the findings of Etok et al. (2012) who stated that large numbers of S. aureus are sensitive to quinolones and aminoglycosides.
Fluoroquinolones (CIP, OFX, and MFX) are bactericidal agents as previously described. Trimethoprim-sulphamethoxazole (SXT) is also a bactericidal agent against S. aureus and inhibits bacterial replication. Aminoglycosides (AK and CN) are the only ribosome-targeting antibiotics that are bactericidal. This is due to their unique mechanism of action in causing misreading of mRNA during translation. Linezolid is a synthetic oxazolidinone class of antimicrobial agent that binds to the ribosome and inhibits microbial protein synthesis. Studies have confirmed that linezolid has good activity against most Gram-positive bacteria, specially against MRSA (Foster 2017). Resistance to penicillin was determined at the rate of 100% due to beta-lactamase production (Rağbetli et al.,2016). Inactivation of oxacillin is due to β-lactamase hyperproduction. These hyperproducers of β-lactamase tend to resist oxacillin through limited hydrolysis of the antibiotic, resulting in a phenotype that, with deference to oxacillin, is called oxacillin-resistant S. aureus (MRSA). A second mechanism of resistance to oxacillin is due to the production of an altered penicillin-binding protein (PBP 2’ or PBP 2a) which facilitates bacterial growth and cell wall synthesis at concentrations of β-lactams inhibitory to native penicillin-binding proteins. Yakha et al.(2015) reported that S. aureus showed high resistance against penicillin and oxacillin. In this study (Table 4), S.a3 and S.a9 isolates showed total resistance against CIP and MXF and OFX.
As illustrated in Fig. 2, the incidence of MDR, XDR, and PDR studied isolates were as follows: out of the 40 bacterial strains, 20 (50%) were MDR, 7 (17.5%) were XDR, and 3 (7.5 %) were PDR. The remaining 10 isolates (25%) were detected to be sensitive to most of the tested antibiotics. In the present study, Fig. 3 shows that two P. aeruginosa strains were MDR, and 5 were XDR, 6 E. coli strains were MDR, and one responded as PDR. Out of the 8 Klebsiellaspp., 4 strains were MDR, 2 XDR, and 2 were PDR. Proteus mirabilis strain reacted as an MDR and finally7/10 isolates of the Gram-positive Staph.spp. were MDR.
WHO, (2014) reports showed that approximately 50% of E. coli, K. pneumoniae, S. aureus, and P. aeruginosa were resistant to the majority of antibiotics, such as cephalosporin. The increasing trend in development of antibiotic resistance could be attributed to frequent, unnecessary, and abuse of antibiotics and longer duration of hospitalization.
The studied plant extracts of M. oleifera and M. recutita showed varied levels of antibacterial activity against antibiotic-sensitive and resistant P. aeruginosa, Klebsiella pp., E. coli, Proteus mirabilis, and Staphylococcus spp. isolates. Water and methanol extracts of both plants represented good activity against most of the sensitive and resistant isolates, whereas ethanol extract of both plants showed a lesser activity against nearly all of the isolates. Aqueous and methanol extract of both plants showed high inhibition zones against the studied bacteria isolates (Table 5).
Moreover, most people who use M. oleifera leaves as a traditional means of treatment of various skin ailments and other diseases make use of water-based extract of the leaf. This agreed with Dike-Ndudim et al.(2016) and Muhuha et al. (2018) who noted that the aqueous extract of M. oleifera leaf possesses significant antimicrobial activity against both Gram-negative and Gram-positive bacterial organisms from wounds, thus signaling its broad spectrum of antibacterial activity. Further chemical composition analysis revealed that the M. oleifera leaf extract with antibacterial activities contains alkaloids, polyphenols, flavonoids, anthraquinones, coumarins, tannins, triterpenes, sterols, saponins, and some other secondary metabolites.
The experiments carried out by Daotam et al.(2016) confirmed that the methanol extract of M. oleifera leaves showed different inhibition patterns against different bacterial strains including E. coli, E. aerogenes, K. pneumoniae and P. aeruginosa. Abdalla and Abdelgadir, (2016) reported that water and methanol extracts of M. chamomilla showed different degrees of antibacterial activities against bacteria, including P. aeruginosa, S. aureus, B. cereus and E. coli.
Conclusion
The application of herbal products for the bio-control of diseases, as a novel emerging alternative to antimicrobial treatments leading to nontoxic and more environmental managing for virulent diseases, is a must.
Due to the rapid development of resistance against chemotherapeutic agents (mostly antibiotics), it has become essential currently to think over some substitute and effective therapeutics like herbs.
This study had the potential value to develop herbal products as antibacterial agents against resistant and susceptible bacteria supporting the significant use of plant extracts in treating wound infections related to bacteria and these active extracts will provide useful information for discovering new compounds with better activity and more effect against resistant (MDR, XDR. and PDR) and susceptible bacteria responsible for wound infections than currently available antibiotic agents.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- CDC:
-
Centre for Disease Control and Prevention
- CLSI:
-
Clinical and Laboratory Standards Institute
- ECDC:
-
European Centre for Disease control
- GNB:
-
Gram-negative bacilli
- GPC:
-
Gram-positive cocci
- MBC:
-
Minimum bactericidal concentration
- MDR:
-
Multidrug-resistant
- MIC:
-
Minimum inhibitory concentration
- PDR:
-
Pan drug-resistant
- WHO:
-
World Health Organization
- XDR:
-
Extensive drug-resistant
References
Abalaka ME, Daniyan SY, Oyeleke SB, Adeyemo SO (2012) The antibacterial evaluation of Moringa oleifera leaf extract on selected bacterial pathogens. J Microbiol Res 2(2):1–4
Abdalla RM, Abdelgadir AE (2016) Antibacterial Activity and Phytochemical Constituents of Cinnamomum verum and Matricaria chamomilla from Sudan. Bio Bulletin 2(2):01–00
Aldred KJ, Kerns RJ, Osheroff N (2014) Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574
Ali AA, Alattar SA (2018) Study the protective effect of Matricaria chamomilla flower extract against the toxicity of Entamoeba histolytica induces liver and renal disfunctions in adult albino male rats. Iraqi J Sci 59(2B):832–838
Aljanaby AAJ, Aljanaby IAJ (2018) Prevalence of aerobic pathogenic bacteria isolated from patients with burn infection and their antimicrobial susceptibility patterns in Al-Najaf City, Iraq- a three-year cross-sectional study. F1000Research 7:1157
Bassetti, M.,Vena, A. Croxatto, A., Righi, E. and Guery, B. (2018). How to manage Pseudomonas aeruginosa infections. Drugs Context, 7:212527.
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Bergey’s Manual of Systematic Bacteriology. (1989). Williams, S. T., Sharpe, M. E. and Holt, J. G. (Ed.), pp 399. The Williams and Wilkins Co. Baltimore.Vol. 4.
Bhalchandra HM, Naik DS, Verma KP (2018) Aerobic Bacterial Profile of Wound Infections and Its Sensitivity Pattern at Tertiary Care Hospital. Int J Curr Microbiol App Sci 7(6):1668–1679
Chandra H, Bishnoi P, Yadav A, Patni B, Mishra AP, Nautiyal AR (2017) Antimicrobial Resistance and the Alternative Resources with Special Emphasis on Plant-Based Antimicrobials-A Review. Plants (Basel, Switzerland) 6(2):16
Cheesbrough, M.(1984). Biochemical testing of microorganisms. Medical laboratory manual for tropical countries. Syndicate of the university of Cambridge and Tropical Health Technology. Microbiology, section 5, chapter 36.
CLSI, (2016). Performance standards for Antimicrobial Susceptibility Testing, 18th Informational Supplement. M100-S18. Wayne, PA: Clinical and Laboratory Standards Institute.
Dan MM, Sarmah P, Vana DR, Dattatreya A (2018) Wound Healing: Concepts and Updates in Herbal Medicine. Int J Med Res Health Sci 7(1):170–181
Daotam JK, Touani FK, Kuete V (2016) Antibacterial and antibiotic-modifying activities of three food plants (Xanthosoma mafaffa Lam., Moringa oleifera(L.) Schott and Passiflora edulis Sims) against multidrug-resistant (MDR) Gram-negative bacteria. BMC Complement Altern Med 16(1):9
Das DC, Sinha NK, Chattopadhyay JC, Das M (2013) Antibacterial activity and photochemical analysis of cardanthera difformis druce leaf extracts from west Bengal, India. J. Phytomedicine (5):446–451
Dike-Ndudim JN, Anyanwu GO, Egbuobi RC, Okorie HM, Udujih HI, Nwosu DC, Okolie NJC (2016) Anti-bacterial and phytochemical potential of Moringa oleifera leaf extracts on some wound and enteric pathogenic bacteria. Eur J Bot Plant Sci Phytol 3(1):50–60
Etok CA, Edem EN, Ochang E (2012) Aetiology and antimicrobial studies of surgical wound infections in University of Uyo Teaching Hospital (UUTH) Uyo, Akwa Ibom State, Nigeria. Niger Open Access Sci Rep 1:1–5
Foster TJ (2017) Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev 41(3):430–449
Gomatheswari SN, Jeyamurugan T (2017) Bacteriological Profile and the Antibiotic Susceptibility Pattern of Microorganisms Isolated from Pus/Wound Swab Isolates in Patients Attending a Tertiary Care Hospital in South India. Int J Curr Microbiol App Sci 6(10):1405–1413
Greenwood D (1989) Antibiotic Sensitivity Testing. In: Antimicrobial Chemotherapy. Oxford University Press, New York, pp 91–100
Hooper DC, Jacoby GA (2015) Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31
Kassam NA, Damian DJ, Kajeguka D, Nyombi B, Kibiki GS (2017) Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a Tertiary Hospital, northern Tanzania. BMC Res Notes 10:757
Magiorakos, A.P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M.E., Giske, C.G., Harbarth, S., Hindler, J.F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D.L., Rice, L.B., Stelling, J., Struelens, M.J., Vatopoulos, A., Weber, J.T. and Monnet, D.L. (2012). Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect,18(3): 268–281
Manikandan C, Amsath A (2013) Antibiotic susceptibility of bacterial strains isolated from wound infection patients in Pattukkottai Tamilnadu India. Int J Curr Microbiol App Sci 2(6):195–203
Mohammed, A., Seid, M. E., Gebrecherkos, T., Tiruneh, M., and Moges, F.(2017). Bacterial Isolates and Their Antimicrobial Susceptibility Patterns of Wound Infections among Inpatients and Outpatients Attending the University of Gondar Referral Hospital, Northwest Ethiopia, Int. J. Microbiol, vol. 2017, Article ID 8953829, 10 pages.
Muhuha AW, Kang’ethe SK, Kirira PG (2018) Antimicrobial Activity of Moringa oleifera, Aloe vera and Warbugia ugandensis on MultiDrug Resistant Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Agents 4:168
Nikolaidis I, Favini-Stabile S, Dessen A (2014) Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci 23(3):243–259
Osariemen IJ, Olown SS, Adevbo E, Omon EE, Victoria O, Imuetinyan EJ, Adesuwa E (2013) Aerobic bacteria associated with diabetic wounds in patients attending clinic in a rural community in Nigeria. Glob Res J Microbiol 3(1):8–12
Pallavali RR, Degati VL, Lomada D, Reddy MC, Durbaka VRP (2017) Isolation and in vitro evaluation of bacteriophages against MDR bacterial isolates from septic wound infections. PLoS ONE 12(7):179–245
Palzkill T (2018) Structural and Mechanistic Basis for Extended-Spectrum Drug-Resistance Mutations in Altering the Specificity of TEM, CTX-M, and KPC β-lactamases. Front In Mol Biosci 5:16
Parija SC (2012) Textbook of Microbiology & Immunology, 2nd edn
Perim MC, Borges JDC, Celeste SRC, Orsolin EDF, Mendes RR, Mendes GO, Ferreira RL, Carreiro SC, Pranchevicius MC (2015) Aerobic bacterial profile and antibiotic resistance in patients with diabetic foot infections. Revista Da Sociedade Brasileira de Medicina Tropical 48:546–554
Rağbetli, C., Parlak, M., Bayram, Y., Guducuoglu, H. and Ceylan, N. (2016). Evaluation of Antimicrobial Resistance in Staphylococcus aureus Isolates by Years. Interdiscip Perspect Infect, vol. 2016, Article ID 9171395, 4 pages.
Rahman, M. M., Islam, M.M., Sheikh, S. A., Sharmin, A., Islam, M. S., Rahman, A. and Alam, M. F. (2009). Antibacterial Activity of Leaf Juice and Extracts of Moringa oleifera Lam. against Some Human Pathogenic Bacteria. CMU Journal, 8(2): 219–228
Saha AK, Nandi S, Dhar P (2017) Spectrum of microbial isolates from wound infections in patients admitted in a Tertiary Care Hospital , Kolkata. MGM J Med Sci 4(1):10–18
Saif MMS, Al-Fakih AA, Hassan MAM (2017) Antibacterial activity of selected plant (Aqueous and methanolic) extracts against some pathogenic bacteria. J Pharmacognosy Phytochemistry 6(6):1929–1935
Shakya AK (2016) Medicinal plants: Future source of new drugs. Int J Herb Med 4(4):59–64
Sultana SH, Mawla N, Kawser SH, Akhtar N, Ali MK (2015) Current Microbial Isolates from Wound Swab and Their Susceptibility Pattern in a Private Medical College Hospital in Dhaka city. Delta Med Coll J 3(1):25–30
Tirado-Torres D, Chan-keb CA, Pérez-Balán RA, Ake-Canché B, Gómez Solano MI, Aragón-Gastélum JL, Gómez-López I, Aguirre-Crespo FJ, López-Ramos MC, Gutiérrez-Alcántara EJ (2019) Antimicrobial activity of Moringa oleifera against multidrug-resistant Staphylococcus aureus isolated from raw milk. Appl Ecol Environ Res 17(1):587–599
Usman JG, Sodipo OA, Sandabe UK (2014) In vitro antimicrobial activity of Cucumis metuliferus E. Mey. Ex. Naudin fruit extracts against Salmonella gallinarum. Int J Phytomed 6(2):268–274
WHO. (2005). National policy on traditional medicine and regulation of herbal medicines: Report of a WHO global survey. World Health Organization, Geneva, Switzerland.
WHO. (2014) Antimicrobial resistance global report on surveillance. 〈http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf〉.
Yakha JK, Sharma AR, Dahal N, Lekhak B, Banjara MR (2015) Antibiotic susceptibility pattern of bacterial isolates causing wound infection among the patients visiting B & B hospital. Nepal J Sci Technol 15(2):91–96
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
NA she made the main contributions to the work design; she was involved in microbiological laboratory testing of samples, revising the scientific data, and approving the final version. SS she revised drafting the clinical aspect of the study. She read the manuscript and gave her final approval to be published. SN: she has been involved in microbiological laboratory testing, microbial isolation and antibiotic sensitivity test of the samples. She was responsible for submission of the article as a corresponding author and the resubmission after making the necessary revision and correction of the manuscript. YA performed the clinical work of the study, including patient examination and obtaining the samples to the microbiology laboratory. She recorded the results and data collection.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Agreement for the study, in addition to the ethical approval, was obtained first from the Medical Research Committee of RIO and Kasr El Aini, Cairo,Egypt. All the patients who were enrolled had submitted an informed consent in advance.
Consent for publication
Not applicable (participants were fully anonymous).
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Atef, N.M., Shanab, S.M., Negm, S.I. et al. Evaluation of antimicrobial activity of some plant extracts against antibiotic susceptible and resistant bacterial strains causing wound infection. Bull Natl Res Cent 43, 144 (2019). https://doi.org/10.1186/s42269-019-0184-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-019-0184-9