Abstract
Background
The tomato leafminer, Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae), is a serious pest of solanaceous plant species, mainly Solanum lycopersicum L. The entomopathogenic fungi, an alternative to chemical insecticides, proved to be an efficient biocontrol agent in reducing pest population density. In the present study, an entomopathogenic fungus, Purpureocillium lilacinum isolated from soil was identified based on the morphological and molecular characteristics and its pathogenicity was tested against target pest.
Results
The micromorphological characters showed variations in growth pattern, shape and colour on different cultural media. For molecular analysis, a phylogenetic tree based on ITS/LSU and ITS/β-tubulin (benA) gene regions was constructed which revealed the isolate (FC18) as P. lilacinum (Thom) Luangsa-ard, Houbraken, Hywel-Jones and Samson (Family: Ophiocordycipitaceae). Further, the pathogenicity of P. lilacinum was tested using different spore concentrations (1 × 108, 1 × 107, 1 × 106, 1 × 105 and 1 × 104 spores/ml) on larval and pupal stages of T. absoluta, which showed a dose-dependent mortality. At the highest concentration (1 × 108 spores/ml), the mean mortality of 92.99, 83.05, 72.0% of second, third and fourth instar was observed, respectively. Pupal mortality also showed significant differences at different spore concentrations.
Conclusion
Obtained results showed that the virulence of the indigenous strain of P. lilacinum on T. absoluta can be utilized in the field suppression of the pest as a potent biocontrol agent.
Similar content being viewed by others
Background
Tuta absoluta (Meyrick), (Lepidoptera: Gelechiidae), is known as South American pinworm, is a multivoltine lepidopteran species and one of the most destructive pests of tomato (Solanum lycopersicum) and other solanaceous plant species (Öztemiz 2013). In India, this invasive pest was first reported at Pune, Maharashtra, in October 2014 and now is found in almost all tomato growing states of India (Sharma and Gavkare 2017). Chemical insecticides are commonly used to control the T. absoluta, but frequent use of chemical insecticides has developed pest resistance (Lietti et al. 2005). The chemical insecticides also lead to toxic effects on human health, environment and reduce the population densities of beneficial insects (Roditakis et al. 2015). Therefore, biological control agents are deployed for eco-friendly pest management (Bukhari et al. 2011).
Entomopathogenic fungi (EPF) are considered as important biocontrol agents having potential to cause mycosis and sometimes kill the insect pests (Shah and Pell 2003). Naturally occurring EPF are one of the best alternatives to harmful chemical insecticides. These fungi infect the host by the attachment of conidia (spores) on the external body surface of insect’s cuticle. After breaching the cuticle, the germinating hyphae of fungal pathogen perforate into the haemocoel of insect body where it colonizes and proliferates throughout the host to form hyphal bodies after the replication (Wanchoo et al. 2009). Upon colonization of the host, the EPF release toxic compounds that often lead to the death of the host (Trienens and Rohlfs 2012). To exploit these fungi as biocontrol agents, selective isolation, confirmation of the identity by combined approaches of morphological and molecular characterization is highly essential (Dunlap et al. 2017).
Recent studies have revealed the pathogenicity of the EPF, Purpureocillium lilacinum (Thom) Luangsa-ard, Houbraken, Hywel-Jones and Samson against nematodes and a few insect pests like thrips, bugs, beetles, aphids and white flies (Goffré and Folgarait 2015). Very little information is available on molecular characterization of the indigenous strain of P. lilacinum and its pathogenicity against T. absoluta. Therefore, present study aimed to isolate and characterize the indigenous entomopathogen to evaluate its efficacy against larvae and pupae of T. absoluta under laboratory conditions.
Methods
Isolation of P. lilacinum from soil sample
The EPF were isolated from the soil using bait method of Zimmerman (1969). Two hundred and fifty grams of soil was collected from a depth on 5 cm from the agricultural field of Savitribai Phule Pune University campus, Pune, India (18.5524°N; 73.8267°E), and brought to the laboratory in plastic bags. Five numbers of second instar larvae of Galleria mellonella L. (Lepidoptera: Pyralidae) were released into 50 g of soil placed in plastic bottles as bait for isolation of P. lilacinum. The soil was incubated at 25 ± 2 °C, and 75% R.H. After 7 days, larvae (of G. mellonella) were examined for infection and the dead larvae were separated from the containers. They were surface sterilized with 1% sodium hypochlorite solution for 2 min and then gently rinsed twice with distilled water (Brownbridge et al. 1993). The cadavers were kept in sterile Petri plates moistened with filter papers and incubated at 25 °C. After seven days of the incubation, cadavers were examined for the appearance of fungal growth. Cadavers with external fungal growth were used for isolation of the entomopathogen and subsequent studies. The pure and identified culture of P. lilacinum was accessioned and preserved in the National Fungal Culture Collection of India (NFCCI; WDCM-932), MACS-Agharkar Research Institute, G.G. Agharkar Road, Pune 411,004, India, under accession number NFCCI 5268.
Morphological characterization
The isolated pure fungus was grown on 2 different culture media, Sabouraud dextrose yeast agar (SDYA; 40 g dextrose, 10 g mycological peptone, 15 g agar, 0.1gm chloramphenicol (pH: 5.6)) and Malt extract agar (MEA; 20 g malt extract, 20 g dextrose, 6 g peptone, 15 g agar, 0.1 g chloramphenicol (pH: 5.6)). After 14 days of incubation of culture plates at 25 ± 2 °C, observations on colony characters, shape, diameter, etc., were recorded. Methuen Handbook of Color (Kornerup and Wanscher 1978) was used for recording colour of the cultured colonies. For microscopic characters, the slide cultures technique was followed. Small amounts of mycelial inoculum were inoculated on a small block cut out of PDA plate and placed in the centre of the cleaned glass slide overlaid by coverslip. After 5–7 days of incubation, fungal growth on the PDA block was observed directly under microscope and observations noted. Lactic acid and lactophenol-cotton blue mounts were prepared separately for the observations of conidial and other morphological characters using Olympus (Model CX-41, Japan) and Carl Zeiss (AXIO Imager 2, Germany) microscopes.
Extraction of genomic DNA for molecular identification
The genomic DNA was extracted from 7-day-old fungal culture grown on PDA medium and harvested by scrapping the fungal mass using the fine spatula. The fungal mass was placed in a 2 ml tube containing a ceramic pestle and 60-80 mg sterile glass beads (425 µM, sigma). Homogenization of fungal mass was done with lysis buffer (100 mM Tris HCl (pH-7.5); 50 Mm EDTA, 3% SDS) using a FastDNA® spin kit as per manufacturer’s instructions (MP Biomedicals GmbH, Germany) at 6 M/S for 60 s. The PCR amplification of fragments containing regions encoding to ITS 1–5.8S nrDNA-ITS 2 (ITS), 28S nrDNA (LSU) and beta-tubulin (benA) was performed by using primers ITS5/ITS4(5′-TCCTCCGCTTATTGATATGC-3′)/(5′GGAAGTAAAAGTCGTAACAAGG-3′) (White et al. 1990), and LROR/LR7 (5′-ACCCGCTGAACTTAAGC-3′)/(5′-TACTACCACCAAGATCT-3′) (Rehner and Samuels 1994) and T10/T22 (5′-ACGATAGGTTCACCTCCAGAC-3′)/(5′-TCTGGATGTTGTTGGGAATCC-3′) (Glass and Donaldson 1995), respectively. The purification of PCR products was done by an Axygen PCR cleanup kit (Axygen Scientific Inc., CA, USA) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The sequencing reactions were run on an ABI 3100 automated DNA sequencer (Applied Biosystems, USA).
Phylogenetic analysis and nucleotide sequence submissions
The reference sequences of ITS/LSU and ITS/β-tubulin obtained were combined and aligned manually, using the text editor option of the Molecular Evolutionary Genetics Analysis (MEGA) Version 7.0 (Kumar et al. 2016) for constructing phylogenetic tree (Tables 1 and 2). The isolated nucleotide sequences obtained after sequencing were deposited in GenBank, National Centre for Biotechnology Information, under the accession number (MT67260-ITS, MT672602-LSU, MT792846-benA).
Laboratory bioassay
Rearing of T. absoluta
The culture of T. absoluta was obtained from the Germplasm Division Conservation and Utilization, ICAR-National Bureau of Agricultural Insect Resources, Bangalore, and reared under laboratory conditions for bioassay studies. Adults were released into large breeding cages with tomato seedlings under controlled conditions at 27 ± 2 °C and 55% R.H. Moths were provided with 10% honey solution as a source of food. Newly laid eggs were collected and reared in plastic jars on the tomato leaves for their growth and development. The different larval instars were collected from rearing jars and used for bioassay studies.
Pathogenicity of P. lilacinum against T. absoluta
Fifteen-day-old fungal cultures were scraped using a sterile loop and transferred into 10 ml of sterile distilled water containing 0.02% Tween 80 (Rombach et al. 1986). The suspension was filtered through sterile muslin cloth, and five different spore concentrations (1 × 108, 1 × 107, 1 × 106, 1 × 105 and 1 × 104 spores/ml) were prepared using a Neubauer haemocytometer. The pathogenicity of different conidial concentrations of P. lilacinum was tested on 2nd, 3rd, 4th instar larvae and pupae of T. absoluta. For each instar, 10 larvae per replication (total 6 replicates) were dipped in different spore concentrations for 10 s and then air-dried under the laminar flow. After drying, the treated larvae were released on tomato seedlings whose stems were wrapped in wet cotton to maintain turgor pressure. The control larvae were treated with distilled water with 0.02% Tween 80. To determine the pupal mortality, about 250 g of soil was sieved and placed in 500 ml plastic container and autoclaved for 20 min at 15 psi. The sterilized soil was moistened with distilled water. Then, spore concentration (1 × 104 to 1 × 108) was poured in the soil samples and pupae were released in the plastic containers having sterile soil. Each treatment was replicated 6 times with 10 pupae per replicate. The number of larvae and pupae succumbing to fungal infection was recorded daily, and till 7 days. The control larvae and pupae were treated with sterile distilled water containing 0.02% Tween 80 and maintained in similar way. The experiment was repeated twice.
Virulence analysis
Mortality rates were calculated according to Abbott’s formula (1952)
where T = dead larvae in treatment, C = dead larvae in control.
R studio software (Version 4.1.2; R Core Team, 2020) was used for analysis of data which included factorial CRD and ANOVA.
Results
Isolation of P. lilacinum and morphological characterization
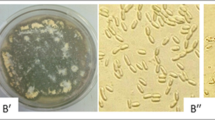
The larvae of G. mellonella placed in the soil samples to recover the fungus were found covered with fungal growth (Fig. 1a). The colony characters of isolated fungus were recorded on MEA medium: colony diameter 50–56 mm, after 14 days of incubation, circular, margins regular, smooth, slightly raised, cottony, dull white (3B9) and reverse yellowish-white (2B5) (Fig. 2a, b). On SYDA medium, the colonies were reddish grey (2B9), 35–40 mm in diameter after 14 days of incubation, cottony, velvety, slightly raised, circular, margins smooth and entire, conidia en masse and reverse greyish yellow (6B4) (Fig. 2c, d).
Colony growth of Purpureocillium lilacinum on different culture media after 14 days of incubation at 25 °C a Frontal view of P. lilacinum on SDYA medium, b Rear view of P. lilacinum on SDYA medium, c Frontal view of P. lilacinum on MEA medium, d Rear view of P. lilacinum on SDYA medium, e, f Microscopic pictures showing phenotypic characteristics, viz. conidiophores arising from mycelial hyphae, phialides, phialospores (conidia) of P. lilacinum
Mycelium simple to branched, septate, smooth walled, bundles of hyphae present. Conidiophores appeared simple to branched arising from superficial hyphae, mono-verticillate to tertiary or quarter verticillate (22–81 × 1.85–2.0 µm). Phialides variable in shape and size, paeciliform, tapered towards apex, (9.25–20.5 × 2.15–2.5 µm), metulae 1–2 in number, straight to curved with a slender neck; conidia mostly fusoid, sub-globose to globose, 2.5–3.15 × 2.0–2.5 µm (Fig. 2e, f). Based on morphological characters recorded, this fungus was identified as Purpureocillium lilacinum (Thom) Luangsa-ard, Houbraken, Hywel-Jones and Samson and the same is deposited and accessioned in NFCCI (5268).
Phylogenetic analysis
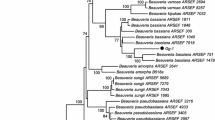
Amplification and sequencing of 2 nuclear and one protein-encoding gene regions (ITS/LSU and ITS/beta-tubulin) were carried out to confirm the isolate’s genotypic characters. The phylogram were generated by combining ITS/LSU and ITS/beta-tubulin with MEGA 7, which includes 31 and 18 nucleotide sequences, respectively. The out-group taxon in both phylograms was Metarhizium marquandii (CBS 182.27) from Clavicipitaceae family (Tables 1, 2). Both ITS/LSU and ITS/beta-tubulin were based on the Tamura 2-parameter model having 917 and 849 sequences positions in the final dataset. Our isolate FC18 clustered with P. lilacinum in both ITS/LSU and ITS/beta-tubulin combined phylogenetic tree with 99 and 97% bootstrap values, respectively. Newly generated sequences of our species were deposited in GenBank. The sequence of 2 genes was aligned and analysed separately by Bayesian and Maximum Likelihood analyses, and the resulting trees were compared (Figs. 3 and 4). There were no conflicts between single gene phylogenies, so the datasets were combined to get phylogram. Nevertheless, both Bayesian and maximum likelihood analyses were useful for discriminating at species level.
Pathogenicity of P. lilacinum against T. absoluta larvae and pupae
Different spore concentrations showed dose-dependent mortality in T. absoluta larvae and pupae. At the highest concentration (1 × 108), the mean mortality was 92.99, 83.05 and 72.0% in second, third and fourth instar larvae, respectively. The corrected mean mortality ranged from 37.53 to 92.83% in second instar, 23.78 to 83.05% in third instar and 11.20 to 72.0% in fourth instar larvae with different conidial concentrations from 1 × 104 to 1 × 108 spores/ml (Table 3). Except to the lower concentration (1 × 104), all the concentrations induced significantly higher mortality (F = 278.23; df = 4; P < 0.001). The mean mortality in instars was observed from 68.15, 56.79 and 41.98% in second, third and fourth instar larvae, respectively (F = 148.75; df = 4; P < 0.001). It was evident from data that with an increase in concentration, the mortality rate was increased. There were significant differences in mortality within a larval instar with increasing conidial concentration (F = 9.52; df = 4; P < 0.001). The LC50 values for the EPF, P. lilacinum, were 5.2 × 107, 4.5 × 106, 3.9 × 105 spores/ml against second, third, and fourth instar larvae, respectively (Table 5).
The corrected mean mortality of pupae was 74.17% at highest concentration 1 × 108 spores/ml, and lowest pupal mortality 11.67% was observed at 1 × 104 spores/ml (Table 4). There was significant difference in all the treatments (F = 23.4; df = 4; P < 0.001). With an increase in concentration, the mortality rate also increased. The pupal mortality from P. lilacinum was dose-dependent. The LC50 value of P. lilacinum for pupae was 3.9 × 105 spores/ml (Table 5). The larvae treated with fungal preparations fed much as lesser than to control larvae. The treated larvae became stiff and hard after death. The white mycelial growth was appeared after 48 h at the highest concentration and after 72 h at the lowest concentration.
Discussion
The phenotypic and genetic variability of EPF plays a crucial role in developing biopesticide because this variability impacts the efficacy of the fungi against many arthropod species (Du et al. 2019). The EPF, P. lilacinum, is a ubiquitous filamentous fungus isolated from soil, decaying vegetation, insects and nematodes (Quandt et al. 2014). In the present study, the morphological characters of P. lilacinum, viz. colony diameter, shape, size, texture and colour, varied on SDYA and MEA media. Other studies have also shown the variation in morphology of P. lilacinum when grown on various media (Nawar et al. 2018), which showed variation in the morphology of the fungus. From present study, it is evident that P. lilacinum may exhibit variations in colour in response to different nutrient media, as shown by Samson (1974). The mycelial structure, conidiophores, phialides and size of conidia are similar to that found in the previous studies (Dyaranthe et al. 2020).
Fungi from the order Hypocreales are complex and difficult to identify by morphological characters and require genomics to decode the complex nature of species identification (Dornburg et al. 2017). Molecular identification by the sequencing of the ITS region has some limitations, so the other genes are necessary to be sequenced for species delimitation (Kredics et al. 2015). In this context, our studies revealed the phylogenetic description of P. lilacinum by amplification of 3 gene sequences including one protein-encoding regions (ITS, LSU and beta -tubulin). The combined ITS/LSU showed that the isolate FC18 belongs to the P. lilacinum of Ophiocordycipitaceae. Similarly, the combined tree of ITS/beta-tubulin revealed the same analysis. Similar studies on phylogenetic analysis were done by Sun et al. (2021).
Many workers reported that EPF have the ability to infect different insect orders (Majeed et al. 2017). The rate of virulence of EPF depends on host range, pattern of virulence factors and level of their expression (Khanday et al. 2018). The pathogenicity of P. lilacinum was tested against different insect orders (Johny et al. 2012). In the present studies, P. lilacinum was tested against second, third and fourth instar T. absoluta larvae and it proved to be pathogenic to all instars at different concentrations. The present data also revealed dose-dependent mortality. It was evident that younger instars were more prone to the fungal infection than the older ones and were in accordance with Spodoptera litura (L.) (Lepidoptera: Noctuidae) (Purwar and Sachan 2005). Similar findings of P. lilacinum were observed in Phthorimaea operculella (Zeller) (Lepidoptera: Gelechiidae) and Leptinotarsa decemlineata (Say) (Coleoptera: Chrysomelidae) when treated with fungal concentration of 108 spores/ml (Kepenecki et al. 2015). Moreover, the infectivity of P. lilacinum was also observed against the cotton aphid, Aphis gossypii (Glover) (Hemiptera: Aphididae) under greenhouse and field conditions (Lopez et al. 2014). The dose-dependent mortality was documented for Acromyrmex lundii (Hymenoptera: Formicidae) (Guerin Meneville) with different P. lilacinum concentrations which reveals the competitive capability of the fungus (Goffré and Folgarait 2015). Highest virulence of P. lilacinum in third and fourth instar larvae of S. litura and Plutella xylostella L. (Lepidoptera: Plutellidae) was demonstrated by Nguyen et al. (2017) which were in concordance with present data. Moreover, the present findings documented the pathogenicity of P. lilacinum concentration against T. absoluta pupae. Schemmer et al. (2016) also reported that EPF were virulent and induced infection to leaf-miner pupae of Cameraria ohridella (Deschka and Dimic) (Lepidoptera: Gracillariidae) under laboratory conditions. Among all isolates tested, Isaria fumosorosea (CO10-IFu) from the same fungal family, Cordycipitaceae, demonstrated promising pathogenic attributes against C. ohridella pupae. The present study is the first report of infection of T. absoluta pupae with P. lilacinum. Obtained data indicate the efficiency of P. lilacinum against pupal stage, but future trials on semi-field and field testing are needed.
In this study, as the concentration of spores increased, the mortality rate of larvae and pupae of T. absoluta increased significantly. The virulence difference of P. lilacinum depends on the application method and the conidial concentrations attached to the cuticle of the studied insect (Rambadan et al. 2011). Further, the previous studies revealed that the range of inoculum has a great significance in causing the infection in the host (Pujol et al. 2011). Other important factors like temperature and humidity, hosts fitness and size also play vital role in causing infection.
Conclusions
In present study, P. lilacinum had shown the potential to infect and kill different larval instars and pupae of T. absoluta with a wide range of variations against different stages of T. absoluta. Morphological and molecular characterizations are useful tools for distinguishing the complexity of EPF. Future investigations are required to evaluate the efficacy of this fungus under field conditions and to determine the most viable route for its application.
Availability of data and materials
Not applicable.
Abbreviations
- MEA:
-
Malt extract agar
- SYDA:
-
Sabouraud dextrose yeast agar
- ITS:
-
Internal transcribed sequence
- LSU:
-
Large subunit
- benA:
-
β-Tubulin
- MEGA 7.0:
-
The Molecular Evolutionary Genetics Analysis Version 7.0
- LC50 :
-
Lethal concentration.
- ANOVA:
-
Analysis of Variance
References
Abbott WS (1952) A method of computing the effectiveness of an insecticide. J Econ Entomol 18:265–267. https://doi.org/10.1093/jee/18.2.265a
Brownbridge M, Humber RA, Parker BL, Skinner L (1993) Fungal entomopathogens recovered from Vermont forest soils. Mycologia 85:358–361. https://doi.org/10.2307/3760695
Bukhari T, Takken W, Koenraadt C (2011) Development of Metarhizium anisopliae and Beauveria bassiana formulations for control of malaria mosquito larvae. Parasit Vectors 4:23. https://doi.org/10.1186/1756-3305-4-23
Dornburg A, Townsend JP, Wang Z (2017) Maximizing power in phylogenetics and phylogenomics: a perspective illuminated by fungal big data. Adv Genet 100:1–47. https://doi.org/10.1016/Bs.Adgen.2017.09.007
Du C, Yang B, Wu J, Ali S (2019) Identification and virulence characterization of two Akanthomyces attenuates isolates against Megalurothrips usitatus (Thysanoptera: Thripidae). Insects 10(6):168. https://doi.org/10.3390/insects10060168
Dunlap CA, Ramirez JL, Mascarin GM, Labeda DP (2017) Entomopathogen ID: a curated sequence resource for entomopathogenic fungi. Antonie Van Leeuwenhoek 111:897–904. https://doi.org/10.1007/s10482-017-0988-2
Dyaranthe MC, Jones EBG, Maharachchikumbura SSN, Devadatha BS, Sarma VV, Khongphinitbunjong K, Chomnunti P, Hyde KD (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11:1–188. https://doi.org/10.1007/s40011-021-01229-y
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved gene from the filamentous Ascomycetes. Appl Environ Microbiol 61:1323–1330. https://doi.org/10.1128/aem.61.4.1323-1330.1995
Goffré D, Folgarait PJ (2015) Purpureocillium lilacinum, potential agent for biological control of the leaf-cutting ant Acromyrmex lundii. J Invertebr Pathol 130:107–115. https://doi.org/10.1016/j.jip.2015.07.008
Johny S, Kyei-Poku G, Gauthier D, Frankenhuyzen KV (2012) Isolation and characterisation of Isaria farinose and Purpureocillium lilacinum associated with emerald ash borer, Agrilus planipennis in Canada. Biocontrol Sci Technol 6:723–732. https://doi.org/10.1080/09583157.2012.677808
Kepenekci I, Oksal E, Saglam HD, Atay T, Tulek A, Evlice E (2015) Identification of Turkish isolate of the entomopathogenic fungi Purpureocillium lilacinum (syn: Paecilomyces lilacinus) and its effect on potato pests, Phthorimaea operculella (Zeller) (Lepidoptera: Gelechidae) and Leptinotarsa decemlineata (Say) (Coleoptera: Chyrosomelidae). Egypt J Biol Pest Control 25:121–127. https://doi.org/10.81043/aperta.78523
Khanday AL, Buhroo AA, Ranjith AP, Mazur S (2018) Laboratory evaluation of entomopathogenic fungi as biological control agents against the bark beetle Pityogenes scitus Blandford (Coleoptera: Curculionidae) in Kashmir. Folia for Pol Ser A-Forestry 60:83–90. https://doi.org/10.2478/ffp-2018-0008
Kornerup A, Wanscher JH (1978) Methuen handbook of color, 3rd edn. Eyre Methuen, London
Kredics L, Narendran V, Shobana CS, Vagvolgyi C, Manikandan P (2015) Indo-Hungarian Fungal Keratitis Working Filamentous fungal infections of the cornea: a global overview of epidemiology and drug sensitivity. Mycoses 58:243–260. https://doi.org/10.1111/myc.12306
Kumar S, Stecher G, Tamura K (2016) MEGA: 7 Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lietti M, Botto E, Alzogaray RA (2005) Insecticide resistance in Argentine populations of Tuta absoluta, (Meyrick) (Lepidoptera: Gelechiidae). Neotrop Entomol 34:113–119. https://doi.org/10.1590/S1519-566X2005000100016
Lopez DC, Zhu-Salzman K, Ek-Ramos MJ, Sword GA (2014) The entomopathogenic fungal endophytes Purpureocillium lilacinum (Formerly Paecilomyces lilacinus) and Beauveria bassiana negatively affect aphid reproduction under both greenhouse and field conditions. PloS one 9:103891. https://doi.org/10.1371/journal.pone.0103891
Majeed MZ, Fiaz M, Ma CS, Afzal M (2017). Entomopathogenicity of three muscardine fungi, Beauveria bassiana, Isaria fumosorosea and Metarhizium anisopliae, against the Asian citrus psyllid Diaphorina citri Kuwayama (Hemiptera: Psyllidae). Egypt J Biol Pest Control 27.
Nawar MA, Abo-Elnasr AA, Kobisi AN, Hefnawy GA (2018) Evaluation of Acaricidal activity of Purpureocillium lilacinum isolated from Egyptian soil against Tetranychus urticae. Egypt J Desert Res 681:57–172. https://doi.org/10.21608/ejdr.2019.8033.1021
Nguyen HC, Tran TV, Ngyuen QL, Nguyen NN, Nguyen MK, Nguyen NTT et al (2017) Newly isolated Paecilomyces lilacinus and Paecilomyces javanicus as novel biocontrol agents for Plutella xylostella and Spodoptera litura. Notu Bot Hort Agrobot Clujnapoca. 45:280–286. https://doi.org/10.15835/nbha45110726
Öztemiz S (2013) Tuta absoluta Povolny (Lepidoptera: Gelechiidae), the exotic pest in Turkey. Rom J Biol Zol 59:47–58
Pujol IA, Ortoneda C, Pastor J, Mayayo E, Guarro J (2011) Experimental pathogenicity of three opportunist Paecilomyces species in a murine model. J Mycol Med 12:86–89
Purwar JP, Sachan GC (2005) Biotoxicity of Beauveria bassiana and Metarhizium anisopliae against Spodoptera litura and Spilartcia oblique. Ann Plant Prot Sci 13:360–368
Quandt CA, Kepler RM, Gams W, Araujo JP, Ban S, Evans HC, Hughes D, Humber R, Hywel-Jones N (2014) Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus 5:121–134. https://doi.org/10.5598/imafungus.2014.05.01.12
Rambadan S, Jugmohan H, Khan A (2011) Pathogenicity and haemolymph protein changes in Edessa meditabunda F. (Hemiptera: Pentatomidae) infected by Paecilomyces lilacinus. J Bio Pest 4:169–175
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for statistical analysis computing, Vienna, Austria. https://www.R-project.org
Rehner SA, Samuels GJ (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634. https://doi.org/10.1016/S0953-7562(09)80409-7
Roditakis E, Grispou M, Nauen R, Vasakis E, Stavrakaki M, Gravouil M, Bassi M (2015) First report of Tuta absoluta resistance to diamide insecticides. J Pest Sci 88:9–16. https://doi.org/10.1007/s10340-015-0643-5
Rombach MC, Aguda RM, Shepard BM, Roberts DW (1986) Entomopathogenic fungi (Deuteromycotina) in the control of the black bug rice, Scotinophara coarctata (Hemiptera: Pentatomidae). J Invertebr Pathol 48:174–179. https://doi.org/10.1016/0022-2011(86)90120-5
Samson RA (1974) Paecilomyces and some allied hyphomycetes. Stud Mycol 6:1–119
Schemmer R, Chladekova P, Medo J, Barta M (2016) Natural prevalence of entomopathogenic fungi in Hibernating pupae of Cameraria ohridella (Lepidoptera: Gracillariidae) and virulence of selected isolates. Plant Protect Sci 52:199–208. https://doi.org/10.17221/110/2015-PPS
Shah PA, Pell JK (2003) Entomopathogenic fungi as biological control agents. Appl Microbial Biotechnol 61:413–423. https://doi.org/10.1007/s00253-003-1240-8
Sharma PL, Gavkare O (2017) New distributional pest Tuta absoluta (Meyrick) in north-western Himalayan region of India. Natl Acad Sci Lett 40:217–220. https://doi.org/10.1127/entomologia/2017/0489
Sun Y, Huang H, Liu Y, Liu S, Xia J, Zhang K, Geng J (2021) Organization and phylogenetic relationships of the mitochondrial genomes of Speiredonia retorta and other lepidopteran insects. Sci Rep 11:2957. https://doi.org/10.1038/s41598-021-82561-1
Trienens M, Rohlfs M (2012) Insect-fungus interference competition-the potential role of global secondary metabolite regulation, pathway-specific mycotoxin expression and formation of oxylipins. Fungal Ecol 5:191–199. https://doi.org/10.1016/j.funeco.2011.07.009
Wanchoo A, Lewis MW, Keyhani NO (2009) Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 155:3121–3133. https://doi.org/10.1099/mic.0.029157-0
White TJ, Burns T, Lee S, Taylor J (1990) Amplification and Direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc Guide Methods Appl 18:315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1
Zimmerman G (1969) The Galleria bait method for detection of entomopathogenic fungi in soil. J Appl Entomol 102:213–215. https://doi.org/10.1111/j.1439-0418.1986.tb00912.x
Acknowledgements
Senior author is thankful to the Savitribai Phule Pune University, Pune, for providing fellowship to conduct research work, and RSP wishes to acknowledge UGC CAS Phase III, DST-PURSE, for the financial assistance. Authors are also thankful to ICAR-National Bureau of Agriculturally Insect Resources, Bengaluru, and Director, MACS-Agharkar Research Institute for providing facilities and assistance throughout this study.
Funding
The funding received by senior author in the form of fellowship not as research grant.
Author information
Authors and Affiliations
Contributions
GKB, RSP and SKS designed the study; RSP contributed to the project coordination. GKB and DKM collected the sample and maintained in the laboratory under optimal conditions and submitted to NFCCI; GKB performed the experiments. GKB and FJW analysed and evaluated the data. GKB wrote the paper. The manuscript was critically evaluated and edited by RSP and SKS. All the authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bali, G.K., Singh, S.K., Maurya, D.K. et al. Morphological and molecular identification of the entomopathogenic fungus Purpureocillium lilacinum and its virulence against Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) larvae and pupae. Egypt J Biol Pest Control 32, 86 (2022). https://doi.org/10.1186/s41938-022-00582-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-022-00582-y