Abstract
Background
Oral squamous cell carcinomas (OSCC) secrete exosomes into the surrounding extracellular environment to promote the horizontal transfer of bioactive molecules including microRNA (miRNA). The primary objective of this study was to explore potential differences in miRNA content between OSCC and OSCC stem cells.
Methods
The OSCC cell lines SCC4, SCC15, SCC25 and CAL27 were used in these studies. The corresponding OSCC stem cells that demonstrated phenotypic adhesion independent tumor spheres (AiTS) were manually isolated. All cells were cultured in DMEM containing 10% exosome-free fetal bovine serum. Exosomes were isolated using Total Exosome Isolation reagent (Invitrogen) and RNA was purified using Total Exosome RNA isolation kit (Invitrogen). Exosome miRNA content was evaluated using miRNA Advanced Taqman Assays for miR-21, − 155, − 133, − 34, − 31, − 32, and − 365. The fold change of miRNA content was calculated using the comparative CT (ΔΔCT) method using miR-16 as an endogenous control.
Results
After successful cell cultures were established, AiTS (cancer stem cells) were manually separated and confirmed using CD133 and Sox-2 biomarkers. Exosomes and extracellular vesicles were successfully isolated from all cell lines and AiTS isolates for miRNA screening. All isolates exhibited miR-16 expression (positive control), but none contained mir-31, − 32, or 133a. Differential expression of miR-21, miR-34 and miR-155 were observed with patterns observed among the cancer cell lines which were distinct from the corresponding AiTS isolates.
Conclusions
Exosomes isolated from these different OSCC stem cell populations displayed nearly consistent downregulation/loss of miR-21 and miR-34 suggesting the possibility of a unique miRNA profile characteristic of oral cancer stem cells. These findings highlight the need to investigate the comprehensive functions of miR-21 and miR-34 in tumor progression and continued research to refine a miRNA profile that could aid in distinguishing tumors with poor prognosis.
Similar content being viewed by others
Background
Oral Cancers include many squamous cell carcinomas that may have varying responsiveness to chemotherapy and radiation treatments [1, 2]. Recent evidence has demonstrated that a sub-population of oral cancers may be cancer stem cells (CSC), which exhibit cancerous but also stem cell phenotypes – including chemotherapeutic and radiation resistance [3, 4]. Based upon these phenotypes, recent efforts have focused on identifying specific biomarkers to identify tumors with sub-populations of CSC [5, 6].
Among the most promising oral cancer biomarkers are circulating microRNAs, which are small, non-coding RNA that are secreted by cancers (and cancer stem cells) into the extracellular environment in small vesicles known as exosomes or oncosomes [7,8,9]. One of the greatest challenges may be the identification of specific microRNAs within these vesicles that could be used for this type of differential diagnosis [10, 11]. For example, many of the hundreds of previously identified microRNAs may be expressed in both cancers and cancer stem cells – although many thousands of microRNAs have yet to be evaluated [12,13,14].
Recent progress has successfully identified several candidate microRNAs that may be differentially expressed among early- versus late-stage oral cancers, which may provide the basis for this type of differential screening [15, 16]. Recent efforts from this group have also contributed to the potential pool of relevant microRNA candidates [17, 18]. More specifically, each of these referenced studies has confirmed the expression of miR-21 and miR-155 in the most aggressive and rapidly dividing oral cancer samples – although none have specifically screened for the expression of these microRNAs among cancer stem cell isolates. In addition, at least two of these studies have found significant correlations with miR-34 and oral cancer prognosis or tumorigenesis – although no studies have screened for the expression of this microRNA among cancer stem cell isolates [15, 16]. Based upon these initial observations, the primary objective of this study was to explore potential differences in specific miRNA content and expression (including miR-21, miR-34, and miR-155) between oral cancers and oral cancer stem cells.
Methods
Cell culture
Three well-characterized oral cancer cell lines SCC4 (CRL-1624), SCC15 (CRL-1623), SCC25 (CRL-1628) and CAL27 (CRL-2095) were obtained from American Tissue Culture Collection (ATCC). SCC4 and SCC15 cells were cultured according to the manufacturer protocol using Dulbecco’s Modified Eagle’s Medium (DMEM):F-12, supplemented with 400 ng/mL hydrocortisone, 10% Fetal Bovine Serum (FBS) and 1% Penicillin-Streptomycin. CAL27 and SCC25 cells were maintained in DMEM with 10% FBS and 1% Penicillin-Streptomycin. All cells were maintained in tissue culture treated flasks (25 cm2) and dishes (6-well) in humidified chambers at 5% CO2.
Isolation of CSC
CSC were isolated from each of the three oral SCC cell lines grown in culture. Each adhesion dependent cell (ADC) line grew into uniform monolayers, revealing small sub-populations of CSC in adhesion-independent tumor spheres (AiDC-TS) developing in cell clusters that were not contact or adhesion-dependent within the monolayer [19]. Separation of these CSC-AiDC-TS was facilitated using a cell scraper to dislodge these clusters and a micropipette to facilitate transfer the AiDC-TS into new cell culture flasks for experimentation [20].
Total cellular RNA isolation
To determine the presence of CSC-specific mRNA biomarkers, RNA was isolated from both ADC and CSC-AiDC-TS using the ABgene Total RNA isolation Reagent kit and the protocol recommended by the manufacturer [18, 20]. Analysis of RNA was accomplished using absorbance readings at 260 nm and 280 nm. RNA quality was measured using the A260:A280 ratio, which should be greater than 1.70. RNA quantity was measured using A260 absorbance reading of 1 = 40 μg/mL RNA, based on the extinction coefficient of RNA in nuclease-free distilled water. Concentrations were determined as 40 x A260 reading multiplied times the dilution factor.
Reverse transcription polymerase chain reaction (RT-PCR)
Expression of CSC-specific mRNA biomarkers was determined using RT-PCR on one ug of total RNA using the ABgene Reverse-iT One-Step RT-PCR kit and a Mastercycler gradient thermocycler using the following primers synthesized by SeqWright:
Control biomarker
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
GAPDH forward primer: 5′-ATCTTCCAGGAGCGAGATCC-3′; 20 nt, 55% GC, 66 °C
GAPDH reverse primer: 5′-ACCACTGACACGTTGGCAGT-3′; 20 nt, 55% GC, 70 °C
Tumor external (surface) biomarker
CD44 forward primer: 5′-GAAAGGCATCTTATGGATGTGC-3′; 22 nt, 45% GC, Tm: 64 °C
CD44 reverse primer: 5′-CTGTAGTGAAACACAACACC-3′; 20 nt; 45% GC, Tm: 61 °C
CSC external (surface) biomarker
CD133 forward primer: 5′-CTCATGCTTGAGAGATCAGGC-3′; 21 nt, 52% GC, Tm: 65 °C
CD133 reverse primer: 5′-CGTTGAGGAAGATGTGCACC-3′; 20 nt, 55% GC, Tm: 66 °C
Tumor internal biomarker
c-myc forward primer: 5′-TCCAGCTTGTACCTGCAGGATCTGA-3′; 25 nt, 52%GC, Tm 72 °C
c-myc reverse primer: 5′-CCTCCAGCAGAAGGTGATCCAGACT-3′; 25 nt, 56%GC, Tm 72 °C
CSC internal biomarker
Sox2 forward primer: 5′-ATGGGCTCTGTGGTCAAGTC-3′; 20 nt, 55% GC, Tm: 67 °C
Sox2 reverse primer: 5′-CCCTCCCAATTCCCTTGTAT-3′; 20 nt, 50% GC, 64 °C
Exosome isolation
Each cell line (ADC and AiDC-TS) was then cultured in media supplemented with exosome-depleted FBS for 24 h prior to exosome isolation. The supernatant (conditioned media) was removed from each tissue culture flask and centrifuged at 2000 x g for 30 min to remove any cells or cellular debris. The cell-free supernatant was then added to Total Exosome Isolation reagent from Life Technology and incubated overnight at 4 °C, as directed by the manufacturer protocol. Exosomes and extracellular vesicles were subsequently isolated using centrifugation at 10,000 x g for 60 min at 4 °C. The exosome-containing pellets were then resuspended in 100 uL of 1X Phosphate-Buffered Saline (PBS).
Exosome RNA isolation
The exosome resuspension was mixed with an equal volume of 2X Denaturing Solution from Life Technology and incubated on ice for 5 min. An equal volume of Phenol:Chloroform was added prior to centrifugation at 10,000 x g for 5 min at 4 °C. The aqueous (upper) phase was removed and mixed 1:1.25 with 100% ethanol (EtOH). This solution was transferred into a filter for subsequent centrifugation at 10,000 x g for 15 s. Each sample-filter was washed with miRNA Wash Solution 1 from life Technology and centrifuged at 10,000 x g for 15 s. This process was repeated with Wash Solution 2/3. Each sample-filter was subsequently placed into a sterile collection tube with 100 uL of heated RNase water and centrifuged for 30 s to collect the exosomal RNA.
Exosome analysis
Exosome isolation was verified using Particle Metrix Nanoparticle Tracking Analysis (NTA) Zeta View software using the manufacturer default setting for extracellular vesicles (EVs) and Nanospheres. Each sample was diluted in sterile 1X PBS to a final concentration of 3.2 × 107 particles/mL to achieve an Average Counted Particles per Frame (ACPF) of 89, well within the recommended particle per frame value range of 40–200. Each measurement involved two cycles scanned at 11 positions with the following settings: Focus – Autofocus; Scattering Intensity – Detected Automatically; Cell temperature – 27.89C sensed. Following data capture, analysis was performed using ZetaView Software 8.05.10 using the following analysis parameters: Max Area: 1000; Min Area: 5; Minimum brightness: 25; Camera 0.712 μm/px.
TaqMan microRNA assays
Analysis of exosomal RNA was accomplished using TaqMan microRNA assays, consisting of Reverse Transcription using10X Reverse Transcription Buffer, 100 mM deoxyribonucleotide triphosphate (dNTP), RNase inhibitor, and MultiScribe Reverse Transcriptase containing three uL of miR specific primer. Settings for the thermocycler were 30 min at 16 °C, 30 min at 42 °C and 5 min at 85 °C, then cooled to 4 °C.
TaqMan small RNA assays were used for quantitative polymerase chain reaction (qPCR) using TaqMan Universal PCR Master Mix II and the product of the reverse transcription reactions. Settings for the thermal cycler were 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles that consisted of 15 s at 95 °C and 60 s at 60 °C. A standard curve was derived from a five-fold serial dilution of cDNA from miR-16, which is used as a positive control for exosomal miRNA and the endogenous reference gene [15, 18].
Statistical analysis
Any differences in miRNA expression were evaluated using two-tailed t-tests isolated from each cell line (ADC and AiDC-TS). Statistical significance level was set at α = 0.05.
Results
Four oral squamous cell carcinoma cell lines (SCC4, SCC15, SCC25 and CAL27) were obtained and cultured to isolate potential CSC from adhesion-independent tumor spheres (Fig. 1). More specifically, as cultures were maintained over time, adhesion-independent tumor spheres (AiTS) developed, which were manually isolated and transferred to separate cell culture flasks for SCC4 (Fig. 1a), SCC15 (Fig. 1b), SCC25 (Fig. 1c) and CAL27 (Fig. 1d) cells. Adhesion dependent (original) and AiTS cultures were then separately cultured and maintained.
In order to determine any differences in mRNA expression and cellular phenotype, RNA was isolated from each of the cell lines and the corresponding AiTS separated from those cell lines (Table 1). These data revealed RNA isolated from each of the original cell cultures had higher concentrations than RNA isolated from the corresponding AiTS for all cell lines. The average RNA concentration from the SCC cell lines was 562 ng/uL (ranging between 278 and 965 ng/uL), which was higher than observed from the AiTS isolated from the corresponding SCC cell lines at 499 ng/uL (ranging between 215 and 892 ng/uL). RNA purity was measured using the ratio of absorbance readings at 260 nm and 280 nm, which revealed an average of 1.68 and 1.53 among the SCC and AiTS isolates, respectively.
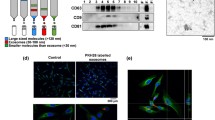
To determine if differences in mRNA expression were evident among the SCC and AiTS isolates, RT-PCR was performed on surface biomarkers to distinguish cancer and cancer stem cells (Fig. 2). These data revealed that the SCC cells (SCC4, SCC15, SCC25 and CAL27) expressed the CD44 biomarker, which may be used to distinguish between normal tissue and oral cancers. This expression was also observed among the AiTS from each cell line, although mRNA expression levels appeared to be slightly lower. However, the expression of CD133, which may be used to distinguish between cancer and cancer stem cells was highly expressed among the AiTS, with much lower (or nearly absent) expression observed among the corresponding cell lines.
RT-PCR screening of Cell Surface Biomarkers. Screening of mRNA isolated from the SCC cell lines and corresponding AiTS isolates revealed higher CD44 (cancer biomarker) mRNA expression among the SCC cell lines than the AiTS isolates. Conversely, CD133 mRNA (cancer stem cell) expression was highest among the AiTS isolates and much lower among the SCC cell lines
To determine if the differential expression of cell surface biomarkers was also correlated with internal biomarkers, the RNA was also screened for c-myc (cancer) and Sox-2 (cancer stem cell) mRNA expression (Fig. 3). These data revealed similar c-myc mRNA expression among both the SCC and AiTS isolates. However, Sox-2 expression was significantly higher among each of the AiTS isolates than was observed among the SCC cell lines.
RT-PCR screening of Internal Biomarkers. Screening of mRNA isolated from the SCC cell lines and corresponding AiTS isolates revealed similar mRNA expression of c-myc among the SCC cell lines and AiTS isolates. However, expression of Sox-2 (cancer stem cell biomarker) was significantly higher among the AiTS isolates than observed among the SCC cell lines
Each SCC cell line and corresponding AiTS isolate was transferred into exosome-free media for subsequent exosome and RNA isolation (Table 2). In brief, following the exosome extraction protocol, RNA was isolated from each of the corresponding exosome preparations, which revealed similar levels of RNA. The average RNA concentration from the SCC exosome preparation (30.8 ng) was not significantly different from the AiTS exosome preparation (30.7 ng), p = 0.9588.
To analyze any potential differences in microRNA expression, TaqMan assays for miR-16, miR-21, miR-34 and miR-155 were used to screen the RNA extracted from the exosomal preparations (Fig. 4). In brief, miR-16 was used to normalize the exosomal microRNA expression, which revealed nearly equivalent amounts of miR-21, miR-34 and miR-155 among the SCC exosomal preparations. However, differential expression was observed among the AiTS. For example, miR-21 expression among the exosomal preparations from the AiTS was down-regulated or absent entirely. Similarly, miR-34 expression was also down-regulated or missing among three of the four AiTS (SCC4, SCC15 and CAL27), with a marked up-regulation observed among the SCC25 AiTS. In addition, miR-155 expression was significantly up-regulated among three of the four AiTS (SCC4, SCC15, and SCC25) with the only down-regulation observed among the CAL27 cells.
TaqMan microRNA assays for exosomal RNA analysis. A. RNA extracted from SCC and AiTS exosomal preparations was screened using TaqMan assays for miR-16, miR-21, miR-34 and miR-155. Expression was normalized to miR-16 (positive control), which revealed similar expression of miR-21, miR-34 and miR-155 among the SCC exosomal preparations. Down-regulation was observed among the AiTS for miR-21 and miR-34 (except SCC25 AiTS). Up-regulation of miR-155 was observed among three of the four AiTS (except CAL27). B. Heat map of microRNA exosomal expression demonstrated differential expression among all AiTS (cancer stem cell) isolates for miR-21, miR-34 and miR-155
To confirm the identification of isolated exosomes, Particle Metrix ZetaView Nanoparticle Tracking Analysis (NTA) was used (Fig. 5). In brief, analysis of exosomal preparations diluted to 3.2 × 107 particles/mL were screened in two cycles and 11 positions to provide verification that the exosomal isolation yielded extracellular vesicles (EVs) and nanoparticles ranging in size between 50 and 200 nm. More specifically, the mean size of EVs from the NTA analysis was 15.9.3 nm +/− 09.5 (STD) with an average volume of 431.6 nm3 +/− 244.9 (STD), which corresponds with known size distributions and parameters for exosomes and extracellular vesicles.
Discussion
The main goal of this study was to examine any potential differences in miRNA content between oral cancers and oral cancer stem cells. The preliminary results of this investigation provide initial evidence that some significant and fairly consistent changes in miRNA expression may be found between oral cancers and their respective stem cell sub-populations, which have been found in other cancers and their corresponding cancer stem cells [21, 22]. This may be consistent with the known functions of microRNAs as regulators of several key tumor-related functions, including angiogenesis [23, 24].
Interestingly, these data may provide evidence that miR-21 might be down-regulated among oral cancer stem cell sub-populations, a key regulator of PI3K/Akt pathway [25, 26]. These observations may be consistent with other studies that suggest control and regulation of the PI3K/STAT3/NF-kB signaling pathway may be more important to tumor functions than critical stem cell or cancer stem cell functions [27, 28]. However, this may be among the first studies to confirm this differential expression between sub-populations derived from the same commercially available oral cancer cell lines.
Another novel finding from this study is the observation that miR-34 may also be down-regulated among the oral cancer stem cell sub-populations. Previous studies have demonstrated that miR-34 may be linked with the regulation of the critical tumor suppressor p53 [29, 30]. As both p53 and miR-34 appear to modulate Wnt signaling, which may also regulate proliferation in both cancers and cancer stem cells – this finding may be consistent with the highly proliferative nature of cancer stem cells [31, 32].
One anomalous finding in this study was miR-34 up-regulation among the SCC25 AiTS, with down-regulation observed among the other three SCC AiTS. More detailed genetic analysis has revealed the presence of underlying dissimilar genetic mutations among these cell lines that may have the potential to influence these experimental outcomes. More specifically, SCC25 cells are known to harbor a homozygous deletion p53 not found in SCC4, SCC15 or CAL27 cells that might influence these findings through disruption of the positive feedback loop between miR-34 and p53 transcriptional regulation [33, 34].
Among the most interesting findings of this study is the observation of miR-155 up-regulation among the AiTS oral cancer stem cell sub-populations. This may be significant as over-expression of miR-155 appears to directly promote cellular proliferation in oral cancers [35,36,37]. These observations may conform with additional evidence that has demonstrated miR-155 may be an indicator of poor prognosis among oral cancers [18, 37, 38].
These findings suggest further studies into these phenomena may be warranted and should include primary tumor explants, which may provide more relevant results than these initial findings from commercially available cell lines - that may also be more readily translated into clinical practice. In addition, another major limitation of this study was the financial constraints on the number of microRNA assays that could be evaluated. Future studies may include additional relevant microRNA candidates, which might greatly enhance the prognostic and diagnostic capabilities of oral cancers.
Conclusions
This study provides preliminary evidence that oral cancers with sub-populations of cancer stem cells may exhibit differential expression of microRNAs. These data may be valuable not only as a prognostic or diagnostic indicator for oral tumors, but also as part of a larger map of potential pathways that may be active or inhibited in these distinct but overlapping cell populations. More research will be needed to determine the clinical and therapeutic relevance of these findings.
Abbreviations
- ADC:
-
Adhesion dependent cell
- AiTS:
-
Adhesion-independent tumor spheres
- ATCC:
-
American Tissue Culture Collection
- CSC:
-
Cancer stem cells
- dNTP:
-
Deoxyribonucleotide triphosphate
- EtOH:
-
Ethanol
- FBS:
-
Fetal bovine serum
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- nt:
-
Nucleotide
- PBS:
-
Phosphate-buffered saline
- RNA:
-
Ribonucleic acid
- SCC:
-
Squamous cell carcinoma
References
Claros MP, Messa CVM, García-Perdomo HA. Adherence to oral pharmacological treatment in cancer patients: systematic review. Oncol Rev. 2019;13(1):402. https://doi.org/10.4081/oncol.2019.402. eCollection 2019 Jan 14 PMID: 31044026.
PDQ Supportive and Palliative Care Editorial Board. Oral Complications of Chemotherapy and Head/Neck Radiation (PDQ®): Patient Version. PDQ Cancer Information Summaries. Bethesda: National Cancer Institute (US); 2002-. 2019. PMID: 26389169
Shin KH, Kim RH. An updated review of Oral Cancer stem cells and their Stemness regulation. Crit Rev Oncog. 2018;23(3–4):189–200. https://doi.org/10.1615/CritRevOncog.2018027501 PMID: 30311574.
Zhang S, Yang X, Wang L, Zhang C. Interplay between inflammatory tumor microenvironment and cancer stem cells. Oncol Lett. 2018;16(1):679–86. https://doi.org/10.3892/ol.2018.8716 Epub 2018 May 16. Review. PMID: 29963133.
Curtarelli RB, Gonçalves JM, Dos Santos LGP, Savi MG, Nör JE, Mezzomo LAM, Rodríguez Cordeiro MM. Expression of Cancer stem cell biomarkers in human head and neck carcinomas: a systematic review. Stem Cell Rev. 2018;14(6):769–84. https://doi.org/10.1007/s12015-018-9839-4 PMID: 30076557.
Mohajertehran F, Sahebkar A, Zare R, Mohtasham N. The promise of stem cell markers in the diagnosis and therapy of epithelial dysplasia and oral squamous cell carcinoma. J Cell Physiol. 2018;233(11):8499–507. https://doi.org/10.1002/jcp.26789 Epub 2018 May 24. Review. PMID: 29797575.
Dharmawardana N, Ooi EH, Woods C, Hussey D. Circulating microRNAs in head and neck cancer: a scoping review of methods. Clin Exp Metastasis. 2019;36(3):291–302. https://doi.org/10.1007/s10585-019-09961-6 Epub 2019 Mar 14. PMID: 30877500.
Mazumder S, Datta S, Ray JG, Chaudhuri K, Chatterjee R. Liquid biopsy: miRNA as a potential biomarker in oral cancer. Cancer Epidemiol. 2019;58:137–45. https://doi.org/10.1016/j.canep.2018.12.008 Epub 2018 Dec 19. Review. PMID: 30579238.
Harrandah AM, Mora RA, Chan EKL. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett. 2018;438:126–32. https://doi.org/10.1016/j.canlet.2018.09.019 Epub 2018 Sep 17. PMID: 30237038.
Olatunji I. Potential application of tumor suppressor microRNAs for targeted therapy in head and neck cancer: a mini-review. Oral Oncol. 2018;87:165–9. https://doi.org/10.1016/j.oraloncology.2018.10.038 Epub 2018 Nov 9. Review. PMID: 30527233.
Cao Y, Green K, Quattlebaum S, Milam B, Lu L, Gao D, He H, Li N, Gao L, Hall F, Whinery M, Handley E, Ma Y, Xu T, Jin F, Xiao J, Wei M, Smith D, Bornstein S, Gross N, Pyeon D, Song J, Lu SL. Methylated genomic loci encoding microRNA as a biomarker panel in tissue and saliva for head and neck squamous cell carcinoma. Clin Epigenetics. 2018;10:43. https://doi.org/10.1186/s13148-018-0470-7 eCollection 2018. PMID: 29636832.
Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, Lo ML. Predictive prognostic value of tissue-based MicroRNA expression in Oral squamous cell carcinoma: a systematic review and meta-analysis. J Dent Res. 2018;97(7):759–66. https://doi.org/10.1177/0022034518762090 Epub 2018 Mar 13. PMID: 29533734.
Irimie AI, Ciocan C, Gulei D, Mehterov N, Atanasov AG, Dudea D, Berindan-Neagoe I. Current Insights into Oral Cancer Epigenetics. Int J Mol Sci. 2018;19(3). doi: https://doi.org/10.3390/ijms19030670. Review. PMID: 29495520
Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and MicroRNAs in Cancer. Int J Mol Sci. 2018;19(2). doi: https://doi.org/10.3390/ijms19020459. Review. PMID: 29401683
Cao M, Zheng L, Liu J, Dobleman T, Hu S, Go VLW, Gao G, Xiao GG. MicroRNAs as effective surrogate biomarkers for early diagnosis of oral cancer. Clin Oral Investig. 2018;22(2):571–81. https://doi.org/10.1007/s00784-017-2317-6 Epub 2018 Jan 3. Review. PMID: 29299731.
Dumache R. Early diagnosis of Oral squamous cell carcinoma by salivary microRNAs. Clin Lab. 2017;63(11):1771–6. https://doi.org/10.7754/Clin.Lab.2017.170607 Review. PMID: 29226639.
Petersen B, Kingsley K. Differential expression of miR-21, miR-133 and miR-155 from exosome fractions isolated from oral squamous cell carcinomas in vitro. J Med Discov. 2016;1(1):jmd6010.
Hunsaker M, Barba G, Kingsley K, Howard KM. Differential MicroRNA Expression of miR-21 and miR-155 within Oral Cancer Extracellular Vesicles in Response to Melatonin. Dent J (Basel). 2019;7(2). doi: https://doi.org/10.3390/dj7020048. PMID: 31052365
Harper LJ, Piper K, Common J, Fortune F, Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36(10):594–603 PMID: 17944752.
Kendall K, Repp MR, Jilka T, Kingsley K. Biomarker screening of oral cancer cell lines revealed sub-populations of CD133-, CD44-, CD24-. And ALDH1- positive cancer stem cells. J Cancer Res Therapy. 2013;1(3):111–8.
Hao J, Zhao S, Zhang Y, Zhao Z, Ye R, Wen J, Li J. Emerging role of microRNAs in cancer and cancer stem cells. J Cell Biochem. 2014;115(4):605–10. https://doi.org/10.1002/jcb.24702 Review. PMID: 24166873.
Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet. 2014;4:295. https://doi.org/10.3389/fgene.2013.00295 Review. PMID: 24427168.
Fanale D, Barraco N, Listì A, Bazan V, Russo A. Non-coding RNAs functioning in colorectal Cancer stem cells. Adv Exp Med Biol. 2016;937:93–108. https://doi.org/10.1007/978-3-319-42059-2_5 Review. PMID: 27573896.
Sheng SR, Wu JS, Tang YL, Liang XH. Long noncoding RNAs: emerging regulators of tumor angiogenesis. Future Oncol. 2017;13(17):1551–62. https://doi.org/10.2217/fon-2017-0149 Epub 2017 May 17. Review. PMID: 28513194.
Sahay S, Tiwari P, Pandey M, Gupta KP. PI3K/Akt pathway and miR-21 are involved in N-ethyl-N-Nitrosourea-induced F1 mouse lung tumorigenesis: effect of inositol Hexaphosphate. J Environ Pathol Toxicol Oncol. 2019;38(1):69–81. https://doi.org/10.1615/JEnvironPatholToxicolOncol.2018026684 PMID: 30806292.
Kowshik J, Nivetha R, Ranjani S, Venkatesan P, Selvamuthukumar S, Veeravarmal V, Nagini S. Astaxanthin inhibits hallmarks of cancer by targeting the PI3K/NF-κΒ/STAT3 signalling axis in oral squamous cell carcinoma models. IUBMB Life. 2019. doi: https://doi.org/10.1002/iub.2104. [Epub ahead of print] PMID: 31251469.
Qiu YF, Wang MX, Meng LN, Zhang R, Wang W. MiR-21 regulates proliferation and apoptosis of oral cancer cells through TNF-α. Eur Rev Med Pharmacol Sci. 2018 ;22(22):7735–7741. doi: https://doi.org/10.26355/eurrev_201811_16395. PMID: 30536317
Bano N, Yadav M, Mohania D, Das BC. The role of NF-κB and miRNA in oral cancer and cancer stem cells with or without HPV16 infection. PLoS One. 2018;13(10):e0205518. https://doi.org/10.1371/journal.pone.0205518 eCollection 2018. PMID: 30372446.
Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle. 2012;11(7):1273–81. https://doi.org/10.4161/cc.19618 Epub 2012 Apr 1. PMID: 22421157.
Kim NH, Cha YH, Kang SE, Lee Y, Lee I, Cha SY, Ryu JK, Na JM, Park C, Yoon HG, Park GJ, Yook JI, Kim HS. p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle. 2013;12(10):1578–87. https://doi.org/10.4161/cc.24739 Epub 2013 Apr 25. PMID: 23624843.
Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, Kang SE, Cha SY, Ryu JK, Na JM, Park C, Kim K, Lee S, Gumbiner BM, Yook JI, Weiss SJ. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011;4(197):ra71. https://doi.org/10.1126/scisignal.2001744 PMID: 22045851.
Brito BL, Lourenço SV, Damascena AS, Kowalski LP, Soares FA, Coutinho-Camillo CM. Expression of stem cell-regulating miRNAs in oral cavity and oropharynx squamous cell carcinoma. J Oral Pathol Med. 2016;45(9):647–54. https://doi.org/10.1111/jop.12424 Epub 2016 Feb 3. PMID: 26841253.
Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35(6):672–88 PMID: 24665023.
Leroy B, Ballinger ML, Baran-Marszak F, Bond GL, Braithwaite A, Concin N, Donehower LA, El-Deiry WS, Fenaux P, Gaidano G, Langerød A, Hellstrom-Lindberg E, Iggo R, Lehmann-Che J, Mai PL, Malkin D, Moll UM, Myers JN, Nichols KE, Pospisilova S, Ashton-Prolla P, Rossi D, Savage SA, Strong LC, Tonin PN, Zeillinger R, Zenz T, Fraumeni JF Jr, Taschner PE, Hainaut P, Soussi T. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res. 2017;77(6):1250–60 Review. PMID: 28254861.
Zeng Q, Tao X, Huang F, Wu T, Wang J, Jiang X, Kuang Z, Cheng B. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int J Mol Med. 2016;37(5):1274–80. https://doi.org/10.3892/ijmm.2016.2529 Epub 2016 Mar 16. PMID: 26986233.
Fu S, Chen HH, Cheng P, Zhang CB, Wu Y. MiR-155 regulates oral squamous cell carcinoma Tca8113 cell proliferation, cycle, and apoptosis via regulating p27Kip1. Eur Rev Med Pharmacol Sci. 2017;21(5):937–44 PMID: 28338203.
Baba O, Hasegawa S, Nagai H, Uchida F, Yamatoji M, Kanno NI, Yamagata K, Sakai S, Yanagawa T, Bukawa H. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J Oral Pathol Med. 2016;45(4):248–55. https://doi.org/10.1111/jop.12351 Epub 2015 Aug 26. PMID: 26307116.
Jakob M, Mattes LM, Küffer S, Unger K, Hess J, Bertlich M, Haubner F, Ihler F, Canis M, Weiss BG, Kitz J. MicroRNA expression patterns in oral squamous cell carcinoma: hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for oral cancer. Head Neck. 2019. doi: https://doi.org/10.1002/hed.25866. [Epub ahead of print] PMID: 31355988.
Acknowledgements
The authors would like to thank the Office of Research at the UNLV School of Dental Medicine and the Graduate and Professional Student Association for their assistance with this project.
Funding
Dr. Katherine M. Howard and Dr. Karl Kingsley are co-investigators on the National Institute of Health (NIH) grant R15DE028431.
Author information
Authors and Affiliations
Contributions
MS, TB, DH and BL performed the experimental protocols and data generation. KK and KMH were responsible for project design and supervision. All authors contributed to this manuscript and have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shoff, M., Booker, T., Leavitt, B. et al. Differential exosome miRNA expression in oral cancer stem cells. ExRNA 2, 3 (2020). https://doi.org/10.1186/s41544-019-0045-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41544-019-0045-6