Abstract
Background
This study aimed to explore the biological activities of miR-330-3p in dextan sulphate sodium (DSS)-induced ulcerative colitis and apoptosis and the direct target of miR-330-3p in this process. HT-29 cells and male C57BL/6 mice were used to examine the function of miR-330-3p in vitro and in vivo, respectively. Expression of miRNA and mRNA was measured using quantitative real time PCR (qRT-PCR). Western blotting was used to measure the change of protein expression. Flow cytometry was used to determine cell apoptosis and luciferase assay was used to confirm the direct target of miR-330-3p.
Results
miR-330-3p expression was increased by DSS in both HT-29 cells and mice. Upregulation miR-330-3p induced cell apoptosis, mice weight loss and ulcerative colitis in vivo, which could prevent by suppression of miR-330-3p. Cell apoptosis related protein expression, cleaved caspase-3 and cleaved PARP was also inhibited by miR-330-3p overexpression and elevated by miR-330-3p inhibition both in vitro and in vivo. Luciferase assay confirmed that 3′ untranslated region (3′-UTR) of XBP1 is the directed target of miR-330-3p and Western blotting results have showed that protein expression of XBP1 was decreased by miR-330-3p mimics and increased by miR-330-3p inhibitor.
Conclusion
miR-330-3p is upregulated by DSS in both HT-29 cells and mice and promoted ulcerative colitis and cell apoptosis by targeting of 3′-UTR of XBP1, which is a key component of ER stress. Inhibition of miR-330-3p prevent DSS-induced ulcerative colitis and cell apoptosis mediated by upregulation of XBP1 expression.
Similar content being viewed by others
Background
Ulcerative colitis is a chronic, inflammatory disorder of the colonic mucosa which can generally extend to the entire colon in a continuous manner [1]. The underlying cause of this disease is still unknown. Many factors have been reported to influence ulcerative colitis, including genetic, environmental and immunological factors [2, 3]. Recent studies have revealed that cellular stress signaling including oxidative stress, mitochondrial hemostasis, autophagy and endoplasmic reticulum (ER) stress contributed to regulation of intestinal epithelial cell function [4].
ER plays a critical role in protein synthesis, folding and modification and calcium storage [5].Misfolded- or unfolded-protein is accumulated in the ER lumen when ER function is dysregulated, which is also called ER stress [6]. Inositol-requiring enzyme 1 (IRE1) is a conserved ER stress sensor and activation of IRE1α can promote mRNA translation to produce X-box–binding protein 1 (XBP1) [7]. XBP1 is a trancription factor, which is related to ER quality control and ER-associated proteolysis [8, 9]. Deletion of XBP1 in mice impaired antimicrobial function and worsened rectal bleeding induced by dextran sulphate sodium (DSS) [10]. When XBP1 was activated by HLJ2, an XBP1 agonist, decreased weight loss, disease activity index (DAI), colon contracture and reduced production of the inflammatory cytokines TNF-α, IL-1β, and IL-6 was observed in mouse model of DSS-induced colitis [11], demonstrating that XBP1 plays a protective role in DSS-induced ulcerative colitis.
microRNAs are a class of small, noncoding RNAs which contain ∼22 nucleotides in length and promote target mRNA cleavage or suppress protein expression by binding the 3′ untranslated region (3′-UTR) of target mRNA [12]. With the development of the in-silico techniques, it is found that single nucleotide polymorphisms is the main cause to induce alternation of miRNAs and their binding sites, resulting in the disease progression [13,14,15]. A number of studies have examined miRNA expression in intestinal tissues and several miRNAs have been identified to participate the pathological process of ulcerative colitis [16]. For example, Upregulation of miR-15 in ulcerative colitis could activate NF-κB signaling pathway through targeting adenosine A2 receptor, which worsened the ulcerative colitis [17]. Overexpression of miR-141 reduced MMP-2 and MMP-9 levels via direct downregulation of CXCL5 expression, indicating a protective effects on ulcerative colitis [18]. Our preliminary data showed that miR-330-3p expression was upregulated in DSS-induced ulcerative colitis, but the mechanism is still unclear. This study aims to explore the pathological function of miR-330-3p in DSS-induced ulcerative colitis and the underlying signaling pathway involved in this process, which will provide a new diagnostic biomarker and a potential therapeutic target for ulcerative colitis.
Results
miR-330-3p was upregulated in DSS-induced ulcerative colitis in mice
After treated with 4% of DSS for 7 days, the body weight of mice was greatly reduced in DSS treated mice compared with mice given normal drinking water (Fig. 1a). DAI score was higher in DSS treated mice than that in control group (Fig. 1b). miR-330-3p expression was dramatically increased in DSS treated mice compared with mice given normal drinking water (Fig. 1c).
miR-330-3p induced cell apoptosis in DSS treated HT-29 cells
In HT-29 cells, miR-330-3p was upregulated by 2% of DSS (Fig. 2a). Cell apoptosis was increased in DSS treated cells and this increase was inhibited in cell transfected with miR-330-3p inhibitor (Fig. 2b and c). The apoptosis related proteins, cleaved caspase-3 and cleaved PARP were determined by Western blotting. The expression of these two proteins was upregulated by DSS. In the cells transfected with miR-330-3p inhibitor, the expression of cleaved caspase-3 and cleaved PARP was reduced compared with the cells treated with DSS (Fig. 2d).
Upregulation of miR-330-3p induced HT-29 cell apoptosis. a miR-330-3p expression was increased by DSS in HT-29 cells; b miR-330-3p inhibitor inhibited cell apoptosis induced by DSS in HT-29 cells; Flow cytometry data shown as two parameter dot-plots; c miR-330-3p inhibitor inhibited cell apoptosis induced by DSS in HT-29 cells; statistically data shown as column; d Upregulation of of cleaved caspase-3 and cleaved PARP expression was inhibited by miR-330-3p inhibitor. ** p < 0.01 vs. Con. or DSS + NC inh; *** p < 0.005 vs. Con. or DSS + NC inh
XBP1 was the direct target of miR-330-3p
TargetScan (http://www.targetscan.org/) was used to predict the potential target of miR-330-3p. The results demonstrated that there was a miR-330-3p binding site in the 3′-UTR of XBP1 (Fig. 3a). Co-transfection of XBP1 3′-UTR and miR-330-3p mimic showed a reduction of luciferase activity compared to negative group control while no significant difference was observed in cells transfected with XBP1 3′-UTR scramble sequence and miR-330-3p mimic (Fig. 3b). The expression of miR-330-3p was elevated by miR-330-3p mimic and repressed by miR-330-3p inhibitor (Fig. 3c). Protein expression of XBP1 was reduced by miR-330-3p mimic and induced by miR-330-3p inhibitor (Fig. 3d).
3′-UTR of XBP1 was the direct target of miR-330-3p. a The putative binding sites between miR-330-3p and 3′-UTR of XBP1; b Luciferase activity was decreased in co-transfection of pGL3-XBP1-WT and miR-330-3p mimic but no change in co-transfection of pGL3-XBP1-MUT and miR-330-3p mimic; c miR-330-3p expression was increased by miR-330-3p mimic and decreased by miR-330-3p inhibitor; d Protein expression was reduce by miR-330-3p mimic and induced by miR-330-3p inhibitor. ** p < 0.01 vs. NC mimic or NC inh; *** p < 0.005 vs. NC mimic or NC inh
Knockdown of miR-330-3p alleviated DSS-induced ulcerative colitis
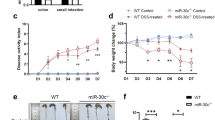
Knockdown of miR-330-3p was achieved by intraperitoneal injection of miR-330-3p antagomir. The body weight was reduced in mice treated with DSS, which was prevented by miR-330-3p antagomir (Fig. 4a). DAI score was increase in mice treated with DSS and this increase was smaller in mice pre-injected with miR-330-3p antagomir (Fig. 4b). Upregulation of miR-330-3p expression was observed in DSS treatment group and injection of miR-330-3p antagomir reduced the miR-330-3p expression compared with negative control group (Fig. 4c). XBP1 expression was reduced in mice treated with DSS (Fig. 4d). miR-330-3p antagomir showed a significant upregulation of XBP1 expression compared with negative control group (Fig. 4d). Both cleaved caspase-3 and cleaved PARP expression was upregulated by DSS in mice and DSS-induced overexpression of cleaved caspase-3 and cleaved PARP reduced by pre-injection of miR-330-3p antagomir (Fig. 4d).
miR-330-3p antagomir alleviated DSS-induced ulcerative colitis in mice. a Body weight loss induced by DSS was prevented by miR-330-3p antagomir; b Elevation of DAI score induced by DSS was decreased by miR-330-3p antagomir; c Upregulation of miR-330-3p was induced by DSS was inhibited by miR-330-3p antagomir; d Change of XBP1, cleaved caspase-3 and cleaved PARP expression induced by DSS was downregulated by miR-330-3p antagomir; ** p < 0.01 vs. Sham or DSS + NC-antagomir; *** p < 0.005 vs. Sham or DSS + NC-antagomir
Discussion
Ulcerative colitis is a kind of inflammatory bowel disease and the common clinical symptoms are: diarrhea, rectal bleeding, abdominal pain and weight loss etc. [19]. Various chemicals have been introduced to induce colitis models in laboratory studies and the most widely used are trinitrobenzene sulfonic acid, oxazolone induced-colitis and DSS [20]. In this study, DSS was used to induce the ulcerative colitis and the results indicated that weight loss and DAI increase was observed, which are typical symptoms of ulcerative colitis. The number of cell apoptosis was also increased in HT-29 cells, an intestinal epithelial cell line. Therefore, this model can be used for further study.
Many studies have revealed that miRNAs were linked to the pathogenesis of colitis [21, 22]. For example, reduction of miR-124 in ulcerative colitis promoted intestinal inflammation mediated by transducer and activator of transcription 3 [23]. Overexpression of miR-31 and miR-155 regulated interleukin-13 signaling pathway via targeting interleukin-13 receptor α-1 in ulcerative colitis [24]. The previous study has found that miR-330-3p was overexpressed in ulcerative colitis [25]. However, no publication has reported the role of miR-330p-3p in ulcerative colitis. Data of the present study revealed that inhibition of miR-330-3p decreased HT-29 cell apoptosis and prevented weight loss and DAI elevation in mouse model, suggesting that suppression of miR-330-3p could ease the symptoms caused by ulcerative colitis, protecting the injury of ulcerative colitis.
As mentioned, the biological function of miRNAs was achieved through bind to 3′-UTR of target mRNA. The 7 nucleotides “seed region”, 2 to 8 at the 5′ end, of miRNA is the key point to target recognition [26, 27]. Target prediction results from TargetScan demonstrated that 3′-UTR of XBP1 bound at 3 to 9 at 5′ end of miR-330-3p and a perfect Watson-Crick match [28] was observed between miRNA and mRNA at the binding site, indicating that 3′-UTR of XBP1 is probably the direct target of miR-330-3p. To confirm this prediction, luciferase assay was conducted [29]. The results of luciferase assay and Western blotting manifested that overexpression of miR-330 reduced XBP1 level. These data validated that miR-330-3p is able to target 3′-UTR of XBP1.
XBP1 is an important transcription factor, which is required for ER stress [30]. Published data have manifested that activation of XBP1 may produce an anti-inflammatory effect [31, 32]. In this study, XBP1 expression was reduced via upregulation of miR-330-3p. Inhibition of miR-330-3p showed a significant upregulation of XBP1 expression and reduction of weight loss and DAI elevation in DSS-induced colitis, which is consistent with the previous finding that activation of XBP1 inhibited the pathogenesis of ulcerative colitis [11]. Therefore, XBP1 could be a therapeutic target for ulcerative colitis.
Conclusion
In conclusion, miR-330-3p was upregulated by DSS in both HT-29 cells and mice and promoted ulcerative colitis and cell apoptosis by targeting of 3′-UTR of XBP1, which is a key component of ER stress. Inhibition of miR-330-3p prevent DSS-induced ulcerative colitis and cell apoptosis and increased XBP1 expression, suggesting that miR-330-3p might be a diagnostic biomarker of ulcerative colitis, and that XBP1 could be a potential therapeutic target for ulcerative colitis. However, all data in the present study are preclinical results, so further clinical study are warranted to figure out the effects of miR-330-3p/XBP1 network in ulcerative colitis.
Methods
Cell culture and miRNA transfection
Human intestinal epithelial cell line, HT-29 cells were purchased from the American Type Culture Collection (ATCC, USA). The cells were cultured with DMEM which was supplemented with heat inactivated fetal bovine serum (FBS; 10%, v/v; Gibco, USA), penicillin (100 U/ml, Gibco, USA), streptomycin (100 μg/ml, Gibco, USA) and incubated under the environment of 5% CO2 at 37 °C. DSS (Sigma-Aldrich, USA) was dissolved in sterile water. HT-29 cells were seeded in sterile 6-well plates. When cell confluence reached to about 80%, cells were treated with 2% of DSS or sterile water for 24 hours (h) [10]. Each experiment was performed in triplicate.
miR-330-3p mimic or its negative control (NC mimic), miR-330-3p inhibitor (inh) or its negative control (NC inh) (Ribobio, China) was dissolved in sterile phosphate-buffered saline (PBS; Jijinchem, China). 70 μl of this solution was gently mixed with 30 μl of Lipofectamine™ 2000 (Invitrogen, USA) and incubated at room temperature for 15–20 minutes (min) to form the complex according to the manufacturer’s instructions. HT-29 cells were seeded in sterile 6-well plates. When cell confluence reached to about 60%, 100 nM of the complex was transfected into cells. After transfection for 24 h, DSS were added and incubated for 24 h as described above. Each experiment was performed in triplicate.
Animal model
C57BL/6 mice (male, 18–20 g, 6–8 weeks) were purchased from Guangdong Medical Laboratory Animal Center and kept under the environment of 23 ± 2 °C, 55 ± 15% humidity, 12 h light/12 h dark cycle. miR-330-3p antagomir or its negative control (NC) was dissolved in sterile PBS and mixed with Lipofectamine™ 2000 (Invitrogen, USA) to form the complex (1 nmol/200 μl of antagomir) as described above. Mice were assigned into 4 groups (n = 8 mice/group) randomly: Group I (Sham): mice were given normal drinking water; Group II (DSS): mice administered with drinking water containing 4% of DSS for 1 week; Group III (DSS + NC-antagomir): mice were administered with 200 μl of NC antagomir mixture by tail vein injection twice per week for 1 week and then administered with drinking water containing 4% of DSS for 1 week; Group IV (DSS + miR-antagomir): mice were administered with 200 μl of miR-330-3p antagomir mixture by intraperitoneal injection twice per week for 1 week and then administered with drinking water containing 4% of DSS for 1 week. DAI was calculated as follows: DAI = (weight loss score + stool characters score + bleeding score)/3 [33]. The mice weights were calculated before killed by overdose of pentobaribital sodium (100 mg/kg) and the colon tissues were collected for further analysis.
All experiment procedures were carried out following the ethical standards under a protocol approved by the Ethics Committee of Shulan(hangzhou) Hospital, and were executed conforming to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (No. 85–23, 1996) [34].
RNA extract and quantitative real time PCR (qRT-PCR) assay
Trizol reagent (Invitrogen, USA) was used to extract total RNA. Reverse transcription was achieved using the PrimeScript RT Master Mix (TaKaRa, Dalian, China) according to the manufacturer’s instruction. The relative RNA expression was examined by qRT-PCR assay using the SYBR Premix Ex Taq II Kit (TaKaRa) on the StepOnePlus system (Applied Biosystems, CA, USA). The data were calculated by means of the 2−ΔΔCt method. The primer sequences (Invitrogen, USA) used in this study were shown as Table 1.
Western blotting
Protein lysates were extracted using RIPA cell lysis buffer (Beyotime, China) and protein concentration was measured using BCA protein assay kit (Beyotime, China). Total protein (50 μg) were electrophoresed on 10% polyacrylamide gel (SDS-PAGE) and transferred to PVDF membranes (Millipore, USA) followed by blockade with 5% of milk for 1 h. Membrans were sequentially probed with the indicated primary antibodies: Cleaved caspase-3 (CST9664, 1:1000 dilution), Cleaved PARP (CST5625, 1:1500 dilution), XBP1 (CST40435, 1:500 dilution) and GAPDH (CST2118, 1:5000 dilution). After incubated with primary antibodies overnight at 4 °C, the membranes were incubated with the appropriate secondary antibodies (Cell Signaling Technology, USA). Finally, the bands were detected using SignalFire™ ECL Reagent (Cell Signaling Technology, USA).
Plasimid and luciferase reporter assay
This protocol was followed the published paper [35]. Briefly, 3′-UTR of XBP1 containing wide type (WT) and scrambled (MUT) miR-330-3p binding sequence were inserted downstream of the firefly luciferase gene in pGL3 to generate the pGL3-XBP1-WT or pGL3-XBP1-MUT plasmid respectively. The constructed plasmids were co-transfected into HT-29 cells with NC mimic or miR-330-3p mimic using Lipofectamine 2000. After 24 h, luciferase activity was assayed using the Luciferase Reporter Assay kit (Abcam, UK) according to the manufacturer’s protocol.
Cell apoptosis by flow cytometry
Flow cytometry was used to detect HT-29 cell apoptosis using an annexin V-FITC apoptosis detection kit (Beyotime Biotechnology) according to the manufacturer’s protocol. The stained samples were analyzed with FACSCalibur (BD, New Jersey, USA). The data of apoptotic cells were shown as two parameter dot-plots.
Statistical analysis
SPSS 10.0 was used to perform the statistical analysis in this study. Student’s t-test was used to compare the difference between two groups. Comparisons of more than two groups was performed using one-factor analysis of variance. All data were demonstrated as means ± standard error of means (S.E.M.). p < 0.05 represents statistically significant.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- qRT-PCR:
-
Quantitative real time PCR
- DSS:
-
Dextran sulphate sodium
- ER:
-
Endoplasmic reticulum
- IRE1:
-
Inositol-requiring enzyme 1
- XBP1:
-
X-box–binding protein 1
- DAI:
-
Disease activity index
- 3′-UTR:
-
3′ untranslated region
References
Danese S, Fiorino G, Peyrin-Biroulet L. Positioning Therapies in Ulcerative Colitis. Clin Gastroenterol Hepatol. 2020;18(6):1280‐1290.e1.
Eisenstein M. Ulcerative colitis: towards remission. Nature. 2018;563(7730):S33.
Yoshimitsu M, et al. The neutrophil/Th1 lymphocyte balance and the therapeutic effect of granulocyte colony-stimulating factor in TNBS-induced colitis of rat strains. J Interf Cytokine Res. 2006;26(5):291–300.
Kaser A, Blumberg RS. Autophagy, microbial sensing, endoplasmic reticulum stress, and epithelial function in inflammatory bowel disease. Gastroenterology. 2011;140(6):1738–47.
So J-S. Roles of endoplasmic reticulum stress in immune responses. Mol Cell. 2018;41(8):705–16.
Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2015;10:173–94.
Cao SS, Kaufman RJ. Unfolded protein response. Curr Biol. 2012;22(16):R622–6.
Olivari S, et al. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J Biol Chem. 2005;280(4):2424–8.
Yamamoto K, et al. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136(3):343–50.
Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–56.
Zhang H, et al. Development of an XBP1 agonist, HLJ2, as a potential therapeutic agent for ulcerative colitis. Eur J Pharm Sci. 2017;109:56–64.
Zhang Y, et al. Emerging roles for microRNAs in diabetic microvascular disease: novel targets for therapy. Endocr Rev. 2017;38(2):145–68.
Kamaraj B, Gopalakrishnan C, Purohit R. In silico analysis of miRNA-mediated gene regulation in OCA and OA genes. Cell Biochem Biophys. 2014;70(3):1923–32.
Bhaumik P, et al. Single nucleotide polymorphisms in microRNA binding sites: implications in colorectal cancer. ScientificWorldJournal. 2014;2014:547154.
Gopalakrishnan C, Kamaraj B, Purohit R. Mutations in microRNA binding sites of CEP genes involved in cancer. Cell Biochem Biophys. 2014;70(3):1933–42.
Schaefer JS. MicroRNAs: how many in inflammatory bowel disease? Curr Opin Gastroenterol. 2016;32(4):258–66.
Zhang H, Li W. microRNA-15 activates NF-κB pathway via down regulating expression of adenosine A2 receptor in ulcerative colitis. Cell Physiol Biochem. 2018;51(4):1932–44.
Cai M, Chen S, Hu W. MicroRNA-141 is involved in ulcerative colitis pathogenesis via aiming at CXCL5. J Interf Cytokine Res. 2017;37(9):415–20.
Ordás I, et al. Ulcerative colitis. Lancet. 2012;380(9853):1606–19.
Randhawa PK, et al. A review on chemical-induced inflammatory bowel disease models in rodents. Korean J Physiol Pharmacol. 2014;18(4):279–88.
Feng Y, et al. MicroRNAs, intestinal inflammatory and tumor. Bioorg Med Chem Lett. 2019;29(16):2051–8.
Cao B, et al. Role of MiRNAs in inflammatory bowel disease. Dig Dis Sci. 2017;62(6):1426–38.
Koukos G, et al. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145(4):842–852.e2.
Gwiggner M, et al. MicroRNA-31 and microRNA-155 are overexpressed in ulcerative colitis and regulate IL-13 signaling by targeting interleukin 13 receptor α-1. Genes. 2018;9(2):85.
Coskun M, et al. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J Gastroenterol. 2013;19(27):4289–99.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Brennecke J, et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85.
Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1(1):18–23.
Clément T, Salone V, Rederstorff M. Dual luciferase gene reporter assays to study miRNA function. In: Rederstorff M, editor. Small non-coding RNAs: methods and protocols. New York: Springer New York; 2015. p. 187–98.
Shaffer AL, et al. XBP1, downstream of blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93.
Ishikawa Y, et al. Spliced XBP1 rescues renal interstitial inflammation due to loss of <em>Sec63</em> in collecting ducts. J Am Soc Nephrol. 2019;30(3):443.
Yap J, et al. TNFα selectively activates the IRE1α/XBP1 endoplasmic reticulum stress pathway in human airway smooth muscle cells. Am J Phys Lung Cell Mol Phys. 2020;318:L483.
Li M-Y, et al. Anti-inflammatory effects of Huangqin decoction on dextran sulfate sodium-induced ulcerative colitis in mice through regulation of the gut microbiota and suppression of the Ras-PI3K-Akt-HIF-1α and NF-κB pathways. Front Pharmacol. 2020;10:1552.
Council, N.R., Guide for the care and use of laboratory animals. 8th 2010. 327(3): p. 963–965.
Xu X-W, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–83.
Acknowledgements
Not applicable.
Funding
None.
Author information
Authors and Affiliations
Contributions
QFC and XMF conceived and designed the experiments, MY and FW analyzed and interpreted the results of the experiments, BX and ZGC performed the experiments. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experiment procedures were carried out following the ethical standards under a protocol approved by the Ethics Committee of Shulan(hangzhou) Hospital, and were executed conforming to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (No. 85–23, 1996).
Consent for publication
Not applicable.
Competing interests
The authors state that there are no conflicts of interest to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, Q., Fang, X., Yao, N. et al. Suppression of miR-330-3p alleviates DSS-induced ulcerative colitis and apoptosis by upregulating the endoplasmic reticulum stress components XBP1. Hereditas 157, 18 (2020). https://doi.org/10.1186/s41065-020-00135-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41065-020-00135-z