Abstract
Background
Cytochrome P450 CYP1A1 helps detoxify the potential carcinogens in tobacco smoke, it was reported that polymorphisms in the coding gene result in variation in the expression and activity levels which alter metabolism and clearance of carcinogens and therefore modify cancer risk. In this work, we aimed to identify CYP1A1 gene polymorphisms associated with lung cancer in Egyptian population and to examine the interaction effect with Tobacco smoking in modulating disease risk.
Methods
A case–control study was conducted on 150 unrelated lung cancer patients and 150 unrelated control subjects. Genomic DNA was extracted and sequencing analysis of CYP1A1 gene was performed on ABI PRISM 3100 genetic analyzer.
Results
Three variants in CYP1A1 gene were identified in heterozygous forms in lung cancer patients I462V, T461N and I286T. A combined variant T461N/ I462V associated with lung cancer and those who carried this variant were 2-times more likely to develop lung cancer (OR = 2.03, 95% CI = 1.81-2.29, P = 0.04), specially the non-small cell type (NSCLC) (OR = 2.20, 95% CI = 1.93–2.50, P = 0.02). Wild type was more frequent among smoker controls (83.3%) compared to smoker lung cancer patients (54.8%), P = 0.03. Association studies to examine the interaction effect of identified variants with Tobacco smoking in modulating disease risk showed no significant associations. Identified polymorphisms showed no significant implication on the stage or the prognosis of the disease.
Conclusion
Our findings support that CYP1A1 polymorphisms play a role in the pathogenesis of lung cancer. In Egyptian population, CYP1A1 I462V, T461N and I286T variants were identified among lung cancer patients and combined T461N/ I462V was a risk variant for NSCLC in non smokers.
Similar content being viewed by others
Background
Lung cancer progression is characterized by cumulative alterations in key molecules involved in the cell cycle, signaling and angiogenesis pathways. Most lung cancer patients demonstrate chromosomal abnormalities at the site of tumor suppressor genes or have mutations in known oncogenes [1].
In the laboratory, chemicals in tobacco reduce the capacity to repair DNA damage in cells from lung cancer patients than cells from normal individuals [2]. As many smokers do not develop lung cancer, it is likely that inherited factors influence the effects of tobacco. There is a considerable interest in variants of genes that help detoxify the carcinogens in tobacco smoke, such as members of the cytochrome P450 (CYP), glutathione S-transferase (GST) and N-acetyltransferase (NAT) gene families [3].
Scientists have identified 57 human CYP genes and 33 pseudo genes divided into 18 families and 42 subfamilies. Now CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5 and other gene polymorphisms have been confirmed and specific metabolic enzyme phenotypes differ with regions and races [4]. CYP gene changes can cause increased activity of the enzyme, decreased activity or even inactivity. In addition, mutations at the substrate recognition sites may lead to changes in enzyme specificity [5].
CYP1A1 plays a major role as a carcinogen activating enzyme within the CYP system. Unlike most CYP enzymes, CYP1A1 expression is mainly found in extra hepatic tissues, including the lung, where it metabolizes and is markedly induced by polycyclic aromatic hydrocarbons (PAHs) [6]. Elevated CYP1A1 inducibility is associated with pulmonary PAH-related DNA adduction [7] and high lung cancer risk [8]. Both CYP1A1 expression and the formation of these PAH-DNA adducts in human lung tissue are highly variable [9–11], possibly due to differing exposure to environmental factors and to genetic polymorphisms affecting the CYP1A1 gene locus [12].
The first variant allele identified was CYP1A1*2A (MspI or m1 polymorphism) and is found in 5% of Caucasians [13]. CYP1A1*2C (Ile462Val or m2 polymorphism) is rare in Caucasians and is usually detected with CYP1A1*2A [14]. The combination of both variants is referred to as CYP1A1*2B. CYP1A1*3, consisting of a T3205C base change (m3), seems to show enhanced enzyme activity, although it is extremely uncommon in Caucasians [13]. Finally, CYP1A1*4, a Thr461Asn (m4) amino acid change, detected in Caucasians with a frequency of roughly 3%, has also been related to greater enzyme catalytic efficiency [15]. These CYP1A1 polymorphisms have been extensively studied with regard to risk of lung cancer. However, whereas some studies report increased risk in the presence of some of the mutations [16, 17], there are many other contradictory results due to ethnic differences [18, 19].
The aim of this work is to identify CYP1A1 gene polymorphisms associated with lung cancer in Egyptian population and to examine the interaction effect with Tobacco smoking in modulating disease risk.
Methods
Subjects
This work was collaboration between National Cancer Institute (NCI) and National Research Center (NRC), Cairo, Egypt. A case–control study was conducted on 150 unrelated adult patients with primary lung cancer and 150 unrelated controls. Patients were presented to NCI from different governorates of Egypt; Cairo, Giza, Qalyubia, Sharqia, Monufia, Kafr El-Sheikh, Minya, Faiyum, Asyut, Sohag and Qena. All subjects included in the study were interviewed to fill a medical questionnaire with special consideration to the lifetime history of tobacco use, residence, occupational history and family history of cancer. Thorough clinical examination and chest radiography were applied. Blood sample was obtained from each subject for sequencing analysis of the human CYP1A1 gene. Sample for histopathology examination of cancer was obtained from each patient either by open biopsy or via bronchoscopy. The exclusion criteria included previous history of cancer, metastasized cancer from other organs, patients with pulmonary fibrosis, acute interstitial pneumonia and previous radiotherapy or chemotherapy or receiving any anti-cancer treatment before enrollment in the study. The study was approved by the ethics committee of the National Research Center. All subjects were aware by the nature of the study and gave a written informed consent.
Methodology
DNA extraction
Genomic DNA was extracted from blood samples obtained from 150 controls and 150 lung cancer patients using QIAamp DNA extraction kit (Qiagen Hilden, Germany, Cat no. 51304) according to the manufacturer's instructions.
Sequence analysis of the CYP1A1 gene
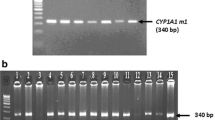
The sequence of the human CYP1A1 gene described in the GenBank (accession number X02612) was used as a reference. The primers used for the amplification and the direct sequencing of all seven exons and exon-intron junctions of the gene are shown in Table 1. PCR for each fragment of CYP1A1 gene was conducted in a 25 μl reaction mixture containing 100 ng of genomic DNA, 20 pmol of each primer and 12.5 μl of AmpliTaq Gold 360 Master Mix (Applied BioSystems, Foster City, CA, USA). PCR cycling conditions consisted of initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30s, annealing at 60 °C for 30s and extension at 72 °C for 30s, followed by a final extension of 5 min at 72 °C. PCR products were checked first on 2% agarose gel for successful amplification (Fig. 1) and was further purified using PureLink Quick PCR Purification Kit (Invitrogen, Germany). The purified PCR products were directly sequenced in both directions using the Big Dye Termination kit (Applied Biosystems, Foster City, CA, USA) and sequences were determined using ABI PRISM 3100 genetic analyzer (Applied Biosystems).
2% agarose gel illustrating the amplification of CYP1A1 gene in two patients. Lanes 1 and 6: amplification of exon 1 (1153-bp). Lanes 2 and 7: amplification of exon 2 (1180-bp). Lane 3 and 8: amplification of exons 3–6 (1233-bp). Lanes 4 and 9: amplification of exon 7a (349-bp). Lanes 5 and 10: amplification of exon 7b (1330-bp). M: Size marker (PhiX174 DNA/HaeIII digest)
Statistical analysis
Data were analyzed using SPSS version 18.0 (Chicago, IL, USA). Data were expressed as number and percentage of total for categorical variables. Chi-square test (χ2) was used to compare the distribution of CYP1A1 genotypes between groups. Likelihood ratio was used when the expected count was less than 5 in more than 20% of the cells. The associations between genotype and risk of lung cancer were estimated by odds ratio (OR) and 95% confidence interval (95% CI) using logistic regression models. The ORs were adjusted for age, smoking status, and pack-years. P-value <0.05 was considered significant.
Results
The study included 300 subjects; 150 unrelated lung cancer patients and 150 unrelated control subjects. A statistical significant age difference was found between controls (mean 43.3 ± 11.1 years) and patients (mean 56.7 ± 9.79 years) (p < 0.001), denoting that older age is associated with higher risk of lung cancer. There was a statistical significant difference in gender between the two groups with 51.3% females and 48.7% males in control group vs. 24% females and 76% males in patients group (X2 = 25.82, P < 0.0001), with 3.5-times increased risk to develop lung cancer in males than in females (OR = 3.49, CI: 2.13–5.69).
Mean pack-years was significantly higher in patients (21.4 ± 2.25) than in controls (4.0 ± 0.77). Smoking habit was significantly higher among patients (X2 = 56.88, P < 0.0001), with an odd ration of 6.69 (CI: 3.99–11.20) denoting a higher risk to develop lung cancer in smokers by 6.7 times the non-smokers.
Our sequencing analysis of CYP1A1 gene identified 2 variants in exon7 in control subjects; CYP1A1*4 1382C > A (T461N) (n = 33) (Fig. 2) and CYP1A1*2C 1384A > G (I462V) (n = 19) (Fig. 3). In lung cancer patients, 3 variants were identified, CYP1A1*2C (n = 17), CYP1A1*4 (n = 46) and combined CYP1A1*2C/ CYP1A1*4 (n = 4). The very low frequent variant in the database CYP1A1 857 T > C (I286T), which was found only 2 times in thousands of human genomes, has been detected in exons 3–6 of the CYP1A1 gene in a non-smoker adenocarcinoma case (Fig. 4). All identified variants were detected in the heterozygous form.
There was no significant difference between controls and lung cancer patients regarding the frequency of identified CYP1A1 variants, except for the combined variants CYP1A1*2C/ CYP1A1*4 which associated with 2-times higher risk of lung cancer (OR = 2.03, 95% C.I:1.81–2.29, P = 0.04), specially the non-small cell type (NSCLC) (OR = 2.20, 95% C.I: 1.93–2.50, P value = 0.02) (Table 2) with no significant difference between the pathological subtypes of NSCLC (P > 0.05).
There was statistical significant difference in the distribution of CYP1A1 variants in relation to smoking habit among lung cancer patients and control subjects. The wild type gene was the most frequent among smoker controls compared to non-smoker controls and lung cancer patients (P = 0.03) (Table 3). Association studies of CYP1A1 polymorphisms and smoking with susceptibility to lung cancer showed no significant association with disease risk (Table 4). No significant association of CYP1A1 polymorphisms and pack year with susceptibility to lung cancer (Table 5). Identified polymorphisms showed no significant implication on the stage or the prognosis of the disease (Tables 6 and 7).
Discussion
CYP1A1 is a polymorphic gene located at 15q24.1 with 7 exons and 6 introns. In addition to the wild-type (CYP1A1*1), 10 variant alleles have been identified [7]. Variants CYP1A1*2A, CYP1A1*2C, CYP1A1*3 and CYP1A1*4 with trivial names m1, m2, m3 and m4; respectively were the most commonly studied for cancer link [20, 21].
About 90% of lung cancer is strongly associated with Tobacco smoking [22]. Our results showed an estimated risk for lung cancer 6.7 times greater in Egyptian smokers than in non-smokers compared to 2.61 times in a previous study [23] and an increased risk to develop lung cancer in males by 3.5-times than in females, which might be attributed to the higher incidence of smoking habit among males.
In our patients, three polymorphisms in CYP1A1 gene were identified; CYP1A1*2C (I462V) was detected in 12.7% of control subjects and 11.4% of patients, and CYP1A1*4 (T461N) identified in 22% of controls and 30.9% of patients. In one non-smoker adenocarcinoma patient, the rare CYP1A1 857 T > C (I286T) was identified. Ethnic difference in the distribution of CYP1A1*2C variant has been demonstrated in lung cancer patients, while few reports are available for CYP1A1*4. Frequencies of CYP1A1*2C ranged from 2.2% to 8.9% in Caucasians and was about 19.8% in Japanese, while CYP1A1*4 allele was found in 2% to 5.7% of a Caucasian population [24–27]. Asian reports rarely observed presence of CYP1A1*4 variant [21, 28, 29], meanwhile, it was more common among whites [30] and it was reported, by itself, as a lung cancer risk factor in Caucasians [12].
CYP1A1*2C and *4 variants have generally been associated with moderate to high risk of lung cancer [21]. These variants are rare in Caucasians and African-Americans. While studies of African-Americans have reported predominantly negative findings [31, 32], studies of Caucasians have been mixed [33, 34]. Although I462V polymorphism is relatively frequent in Asian populations (18% to 25%), the Val allele is rare in Caucasian control populations, occurring in 7% to 13% of people [35–37] which agreed with obtained results in our control population (12.7%).
In the studied Egyptian population, combined variant T461N/ I462V (m2/m4) was found in 2.7% of lung cancer patients associated with 2-times higher risk to develop lung cancer compared to either control or individual m2 or m4 variant carriers.
In an Australian study to identify lung cancer-risk modifying CYP1A1 haplotypes, *2A and *2C variants were significantly over-represented in NSCLC cases compared to controls, whereas *4 variant was under-represented. CYP1A1 haplotypes (in allele order CYP1A1*4, *2C, *2A); CGC and CG associated with increased risk of lung cancer confirming CYP1A1 polymorphisms as minor risk factor for NSCLC. It was reported that CYP1A1*2C increase the overall risk of NSCLC with an odd ratio of 2.88 (95% CI = 1.70–5.00, p < 0.001) [38], in accordance with our results in which CYP1A1*4 and *2C associated with increased risk of NSCLC by 2 times. On the other hand, San Jose et al. reported that I462V and T461V increase the risk to lung cancer in Spanish population, especially to SCLC [12]. Hung et al. found that I462V associated with higher risk for lung cancer, especially for lung adenocarcinoma [39]. Song et al. noted significantly higher risk of lung cancer for I462V variant allele, even in the heterozygous form. However, this elevated risk was restricted to squamous cell carcinoma only, not for adenocarcinoma or other histological types of lung cancer [21]. In our study, no significant difference of lung cancer risk for variant alleles between different pathological subtypes of NSCLC. We could not demonstrate a role of CYP1A1on the stage or the prognosis of the disease, in agreement with a previous study in Taiwan [40].
Controversial results were obtained from previous studies on interaction of smoking and CYP1A1 variants. In the present study CYP1A1*1, wild type, was statistically more frequent among smoker controls. There was no association demonstrated between the heterozygous alleles of CYP1A1 variants, smoking or smoking heaviness in prediction of lung cancer, 75% (3 of 4) of CYP1A1 combined variant (T461N/ I462V) carriers were non-smokers.
Smoking was identified as a predominant risk factor and CYP1A1*2A polymorphism significantly associated with increased lung cancer risk (OR = 1.69; 95% CI = 1.11–2.59, p = 0.01), whereas CYP1A1*2A and *2C and Ile105Val imparted increased risk in non-smokers only [41]. Though, Wenzlaff et al. found no significant association with any of CYP1A1 variants in never smokers [42], a recent pooled analysis by Hung et al. reported more than 2-fold increase in lung cancer susceptibility for both CYP1A1*2B and *2C variants among non-smoker Caucasians [39]. According to Song et al., non-smokers with CYP1A1*2A variant had elevated risk than those homozygous for wild type alleles [21]. Previous study in life-time non-smoking Chinese women reported an elevated risk of lung cancer for both CYP1A1*2B and *2C homozygous genotypes, furthermore, lung cancer risk associated with both polymorphisms was higher in women with lower environmental tobacco smoke exposure [43]. I462V polymorphism is not related to lung cancer overall, but it might play a role at lower levels of Tobacco smoking among subjects with impaired carcinogen detoxification [44].
Conclusion
Beside the wild type CYP1A1*1, three variants of CYP1A1 gene were identified in Egyptian population; CYP1A1*2C, CYP1A1*4 and the rare CYP1A1 c.857 T > C (I286T). Combined variant CYP1A1*2C/ CYP1A1*4 associated with higher risk of lung cancer specially NSCLC among non-smokers. Identification of these variants may help in risk assessment, early detection and improvement of current treatment options for lung cancer patients. Further studies to clarify the role of these variants in the pathogenesis of the disease are needed.
Abbreviations
- Asn:
-
Asparagines
- CYP:
-
Cytochrome P450
- GST:
-
Glutathione S-transferase
- Ile:
-
Isoleucine
- NAT:
-
N-acetyltransferase
- NSCLC:
-
Non-small cell lung cancer
- PAH:
-
Polycyclic aromatic hydrocarbons
- SCLC:
-
Small cell lung cancer
- Thr:
-
Threonine
- Val:
-
Valine
References
Soria JC, Kim ES, Fayette J, Lantuejoul S, Deutsch E, Hong WK. Chemoprevention of lung cancer. Lancet Oncol. 2003;4(11):659.
Breuer RH, Postmus PE, Smit EF. Molecular pathology of non-small-cell lung cancer. Respiration. 2005;72(3):313–30.
Bouchardy C, Benhamou S, Jourenkova N, Dayer P, Hirvonen A. Metabolic genetic polymorphisms and susceptibility to lung cancer. Lung Cancer. 2001;32(2):109–12.
Rodriguez A, Ingelman SM. Cytochrome P450 pharmacogenetics and cancer. J Oncogene. 2006;25(11):1679–91.
Lizasa T, Baba M, Saitoh Y, Suzuki M, Haga Y, Lyoda A, Chang H, Hiroshima K, Itoga S, Tomonaga T, Nomura F, Fujisawa T. polymorphism in the 5 'flanking region of the CYP2E1 gene and elevated lung adenocarcinoma risk in a Japanese population. J Oncol Rep. 2005;14(4):919–23.
Shimada T, Yun CH, Yamazaki H, Gautier JC, Beaune PH, Guengerich FP. Characterization of human lung microsomal cytochrome P-450 1A1 and its role in the oxidation of chemical carcinogens. Mol Pharmacol. 1992;41(5):856–64.
Bartsch H, Petruzzelli S, De Flora S, Hietanen E, Camus AM, Castegnaro M, Alexandrov K, Rojas M, Saracci R, Giuntini C. Carcinogen metabolism in human lung tissues and the effect of tobacco smoking: results from a case–control multicenter study on lung cancer patients. Environ Health Perspect. 1992;98:119–24.
Kouri RE, McKinney CE, Slomiany DJ, Snodgrass DR, Wray NP, McLemore TL. Positive correlation between high aryl hydrocarbon hydroxylase activity and primary lung cancer as analyzed in cryopreserved lymphocytes. Cancer Res. 1982;42(12):5030–7.
Cuzick J, Routledge MN, Jenkins D, Garner RC. DNA adducts in different tissues of smokers and non-smokers. Int J Cancer. 1990;45(4):673–8.
Phillips DH, Schoket B, Hewer A, Bailey E, Kostic S, Vincze I. Influence of cigarette smoking on the levels of DNA adducts in human bronchial epithelium and white blood cells. Int J Cancer. 1990;46(4):569–75.
Anttila S, Vainio H, Hietanen E, Camus AM, Malaveille C, Brun G, Husgafvel-Pursiainen K, Heikkila L, Karjalainen A, Bartsch H. Immunohistochemical detection of pulmonary cytochrome P450IA and metabolic activities associated with P450IA1 and P450IA2 isozymes in lung cancer patients. Environ Health Perspect. 1992;98:179–82.
San Jose C, Cabanillas A, Benitez J, Carrillo JA, Jimenez M, Gervasini G. CYP1A1 gene polymorphisms increase lung cancer risk in a high-incidence region of Spain: a case control study. BMC Cancer. 2010;10:463.
Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, Baranova H, Bathum L, Benhamou S, Boffetta P, Bouchardy C, Breskvar K, Brockmoller J, Cascorbi I, Clapper ML, Coutelle C, Daly A, Dell'Omo M, Dolzan V, Dresler CM. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1239–48.
Georgiadis P, Topinka J, Vlachodimitropoulos D, Stoikidou M, Gioka M, Stephanou G, Autrup H, Demopoulos NA, Katsouyanni K, Sram R, Kyrtopoulos SA. Interactions between CYP1A1 polymorphisms and exposure to environmental tobacco smoke in the modulation of lymphocyte bulky DNA adducts and chromosomal aberrations. Carcinogenesis. 2005;26(1):93–101.
Schwarz D, Kisselev P, Cascorbi I, Schunck WH, Roots I. Differential metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8 -dihydrodiol by human CYP1A1 variants. Carcinogenesis. 2001;22(3):453–9.
Wright CM, Larsen JE, Colosimo ML, Barr JJ, Chen L, McLachlan RE, Yang IA, Wynder EL, Hoffmann D. Smoking and lung cancer: scientific challenges and opportunities. Cancer Res. 1994;54:5284–95.
Lee KM, Kang D, Clapper ML, Ingelman-Sundberg M, Ono-Kihara M, Kiyohara C, Min S, Lan Q, Le Marchand L, Lin P, Lung ML, Pinarbasi H, Pisani P, Srivatanakul P, Seow A, Sugimura H, Tokudome S, Yokota J, Taioli E. CYP1A1, GSTM1, and GSTT1 polymorphisms, smoking, and lung cancer risk in a pooled analysis among Asian populations. Cancer Epidemiol Biomarkers Prev. 2008;17(5):1120–6.
Alexandrie AK, Nyberg F, Warholm M, Rannug A. Influence of CYP1A1, GSTM1, GSTT1, and NQO1 genotypes and cumulative smoking dose on lung cancer risk in a Swedish population. Cancer Epidemiol Biomarkers Prev. 2004;13(6):908–14.
Pisani P, Srivatanakul P, Randerson-Moor J, Vipasrinimit S, Lalitwongsa S, Rautio A, Kraul H, Kojo A, Salmela E, Pelkonen O. Interindividual variability of coumarin 7-hydroxylation in healthy volunteers. Pharmacogenetics. 1992;2:227–33.
Bag A, Jyala NS, Bag N. Cytochrome P450 1A1 genetic polymorphisms as cancer biomarkers. Indian J Cancer. 2015;52:479–89.
Song N, Tan W, Xing D, Lin D. CYP 1A1 polymorphism and risk of lung cancer in relation to tobacco smoking: A case- control study in China. Carcinogenesis. 2001;22:11–6.
Stavridse JC. Lung carcinogenesis: pivotal role of metals in tobacco smoke. Free Radic Biol Med. 2006;41:1017–30.
Ezzeldin N, El-Lebedy D, Darwish A, Ahmed El-Bastawissy A, Shalaby A. Complement factor H polymorphism rs1061170 and the effect of cigarette smoking on the risk of lung cancer. Contemp Oncol (Pozn). 2015;19(6):441–5.
Kawajiri K, Nakachi K, Imai K, Watanabe J, Hayashi S. The CYP1A1 gene and cancer susceptibility. Crit Rev Oncol Hematol. 1993;14:77–87.
Cascorbi I, Brockmoller J, Roots I. A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res. 1996;56:4965–9.
Mrozikiewicz PM, Cascorbi I, Brockmoller J, Roots I. CYP1A1 mutations 4887A, 4889G, 5639C and 6235C in the Polish population and their allelic linkage, determined by peptide nucleic acid-mediated PCR clamping. Pharmacogenetics. 1997;7:303–7.
Aynacioglu AS, Cascorbi I, Mrozikiewicz PM, Roots I. High frequency of CYP1A1 mutations in a Turkish population. Arch Toxicol. 1998;72:215–8.
Duell EJ, Holly EA, Bracci PM, Liu M, Wiencke JK, Kelsey KT. A population-based, case–control study of polymorphisms in carcinogen-metabolizing genes, smoking, and pancreatic adenocarcinoma risk. J Natl Cancer Inst. 2002;94:297–306.
Shaffi SM, Shah MA, Bhat IA, Koul P, Ahmad SN, Siddiqi MA. CYP1A1 polymorphisms and risk of lung cancer in the ethnic Kashmiri population. Asian Pac J Cancer Prev. 2009;10:651–6.
Li Y, Millikan RC, Bell DA, Cui L, Tse CK, Newman B, Conway K. Polychlorinated biphenyls, cytochrome P450 1A1 (CYP1A1) polymorphisms, and breast cancer risk among African American women and white women in North Carolina: A population-based case–control study. Breast Cancer Res. 2005;7:R12–8.
Ishibe N, Wiencke JK, Zuo ZF, McMillan A, Spitz M, Kelsey KT. Susceptibility to lung cancer in light smokers associated with CYP1A1 polymorphisms in Mexican- and African-Americans. Cancer Epidemiol Biomarkers Prev. 1997;6:1075–80.
Taioli E, Ford J, Trachman J, Li Y, Demopoulos R, Garte S. Lung cancer risk and CYP1A1 genotype in African Americans. Carcinogenesis. 1998;19:813–7.
Garcia-Closas M, Kelsey KT, Wiencke JK, Xu X, Wain JC, Christiani DC. A case–control study of cytochrome P450 1A1, glutathione S-transferase M1, cigarette smoking and lung cancer susceptibility (Massachusetts, United States). Cancer Causes Control. 1997;8:544–53.
Dresler CM, Fratelli C, Babb J, Everley L, Evans AA, Clapper ML. Gender differences in genetic susceptibility for lung cancer. Lung Cancer. 2000;30:153–60.
Inoue K, Asao T, Shimada T. Ethnic-related differences in the frequency distribution of genetic polymorphisms in the CYP1A1 and CYP1B1 genes in Japanese and Caucasian populations. Xenobiotica. 2000;30:285–95.
Yang XR, Wacholder S, Xu Z, Dean M, Clark V, Gold B, Brown LM, Stone BJ, Fraumeni Jr JF, Caporaso NE. CYP1A1 and GSTM1 polymorphisms in relation to lung cancer risk in Chinese women. Cancer Lett. 2004;214:197–204.
Chowbay B, Zhou S, Lee EJ. An interethnic comparison of polymorphisms of the genes encoding drug-metabolizing enzymes and drug transporters: experience in Singapore. Drug Metab Rev. 2005;37:327–78.
Larsen JE, ColosimoML YIA, Bowman R, Zimmerman PV, Fong KM. CYP1A1 Ile462Val and MPO G-463A interact to increase risk of adenocarcinoma but not squamous cell carcinoma of the lung. Carcinogenesis. 2006;27:525–32.
Hung RJ, Boffetta P, Brockmöller J, Butkiewicz D, Cascorbi I, Clapper ML, Garte S, Haugen A, Hirvonen A, Anttila S, Kalina I, Le Marchand L, London SJ, Rannug A, Romkes M, Salagovic J, Schoket B, Gaspari L, Taioli E. CYP1A1 and GSTM1 genetic polymorphisms and lung cancer risk in Caucasian non-smokers: a pooled analysis. Carcinogenesis. 2003;24:875–82.
Wang YC, Chen CY, Wang HJ, Chen SK, Chang YY, Lin P. Influence of polymorphism at p53, CYP1A1 and GSTM1 loci on p53 mutation and association of p53 mutation with prognosis in lung cancer. Zhonghua Yi Xue Za Zhi (Taipei). 1999;62(7):402–10.
Ihsan R, Chauhan PS, Mishra AK, Yadav DS, Kaushal M, Sharma JD, Zomawia E, Verma Y, Kapur S, Saxena S. Multiple Analytical Approaches Reveal Distinct Gene-Environment Interactions in Smokers and Non Smokers in Lung Cancer. PLoS ONE. 2011;6(12):e29431.
Wenzlaff AS, Cote ML, Bock CH, Land SJ, Santer SK, Schwartz DR, Schwartz AG. CYP1A1 and CYP1B1 polymorphisms and risk of lung cancer among never smokers: a population-based study. Carcinogenesis. 2005;26(12):2207–12.
Ng DP, Tan KW, Zhao B, Seow A. CYP1A1 polymorphisms and risk of lung cancer in non-smoking Chinese women: Influence of environmental tobacco smoke exposure and GSTM1/T1 genetic variation. Cancer Causes Control. 2005;16:399–405.
London SJ, Yuan JM, Coetzee GA, Gao YT, Ross RK, Yu MC. CYP1A1 I462V genetic polymorphism and lung cancer risk in a cohort of men in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2000;9(9):987–91.
Acknowledgements
Authors would like to thank STDF for the financial support of this work.
Funding
This work was funded by a grant from the Science and Technology Development Fund of Egypt (STDF), project ID 5016.
Availability of data and material
Not applicable.
Authors’ contributions
NE designed the study, AD and AE collected the samples, DE, MH, SA and MA performed the molecular analysis, AS analyzed the data, NE and DE wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the ethics committee of National Research Center of Egypt and informed consent was obtained from all participants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ezzeldin, N., El-Lebedy, D., Darwish, A. et al. Genetic polymorphisms of human cytochrome P450 CYP1A1 in an Egyptian population and tobacco-induced lung cancer. Genes and Environ 39, 7 (2017). https://doi.org/10.1186/s41021-016-0066-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41021-016-0066-4