Abstract
Introduction
This study determined the association of TLR4 Asp299Gly and Thr399Ile with uncomplicated and severe malaria among Nigerian children of similar ethnic background in Lagos. The association of these SNPs with high parasite density, malnutrition, hyperpyrexia and anaemia was also investigated.
Methods
Genomic DNA of the study participants was screened for the genotypes of TLR4 Asp299Gly and Thr399Ile by PCR-RFLP. Anthropometric measurement was performed on the Pf infected children stratified into asymptomatic malaria (control), uncomplicated and severe malaria (case). Parasites were detected by light microscopy and Hardy Weinberg Equilibrium (HWE) of SNP genotypes was also determined.
Results
A total of 279 children comprising 182 children (62.1 % male; mean ± SEM age, 57.3 ± 1.7 months) with clinical falciparum malaria and 97 children (55.7 % male; mean ± SEM age, 55.6 ± 2.5 years) with asymptomatic falciparum malaria were enrolled. All the genotypes of both TLR4 SNPs were found in the study population with their minor alleles: 299Gly and 399Ile, found to be 17.6 % and 14.7 % in severe malaria children. Unlike in asymptomatic population, the genotype distribution of TLR4 Asp299Gly SNP was not in HWE in the clinical malaria group but did not condition susceptibility. However, Asp299Gly and Thr399Ile polymorphisms were found to increase the risk of severe malaria 3-fold and 8-fold respectively (P < 0.05). They also increased the risk of severe anaemia, high parasite density and severe malnutrition 3.8 -5.3-fold, 3.3 – 4.4-fold and 4-fold respectively.
Conclusions
Based on the above findings, we conclude that TLR4 Asp299Gly and Thr399Ile polymorphisms may modulate susceptibility to severe malaria among Nigerian children of Yoruba ethnic background.
Similar content being viewed by others
Introduction
Despite a reduction by 54 % in the African region since 2000, malaria due to Plasmodium falciparum still accounts for 18 % of child deaths [1, 2]. In 2012, 460,000 deaths out of the estimated 627,000 global malaria deaths were reported for children below 5 years with about 86 % of these deaths occurring in sub-Saharan Africa [2]. However, severe malaria occurs in less than 5 % of children below 5 years and this has been attributed to the host genetic factors. [2, 3]. Malaria is an inflammatory disease, which is initiated through recognition of parasite toxin such as glycophosphatidyl inositol (GPI) by innate immune cells such as monocytes, dendritic and macrophages for activation and elicitation of intracellular signal transduction pathway for the production of pro-inflammatory cytokines such tumor necrosis factor-alpha (TNF-α), interleukin-12 (IL-12) and interferon gamma (IFN-γ) [4, 5]. Timely and appropriate production of these cytokines in the early phase of infection and their down regulation by anti-inflammatory cytokines such as IL-10 in the later phase of infection has been shown to be crucial for parasitaemia control and avertion of host tissue damage and severe malaria [4, 5]. These events have been observed in infected African children who are non-susceptible compared to severe malaria susceptible children [6, 7]. Contrastingly, innate immunity dyregulation has been associated with risk factors and syndromes of severe malaria, including malnutrition, severe anaemia hyperparasitaemia or high parasite density [8, 9].
Toll-like receptor 4, a major pathogen recognition receptor (PRR) expressed on membrane surface of innate immune cells is genetically encoded by the TLR4 gene (GeneID = 7099; 9q33.1), which spans a genomic region of ~13.3 kb with three exons (NCBI; http://www.ncbi.nlm.nih.gov/) [10] This PRR is a well-established receptor for the toxigenic GPI of P. falciparum and Trypanosoma cruzi as well as for lipopolysaccharide of gram negative bacteria and other pathogen associated molecular patterns (PAMPS) in gram positive bacteria, fungi and viruses [10–12]. Meanwhile, in vitro and animal model have shown the two most-studied non-synonymous single nucleotide polymorphisms (SNPs) of TLR4: an A to G transition (SNP ID = rs4986790), resulting in Aspartate-Glycine substitution at position 299 (TLR4Asp299Gly) and C to T transition (SNP ID = rs4986791), resulting in threonine-isoleucine substitution at position 399 (TLR4Thr399Ile), to cause altered GPI binding, 50 % reduction in TLR4 expression on the membrane surface of innate immune cells, induce LPS hyporesponsiveness and excessive production of pro-inflammatory cytokines [11, 13, 14]. Subsequent case–control studies then established associations TLR4 Asp299Gly or Thr399Ile polymorphism with death from septic shock and susceptibility to typhoid fever, tuberculosis, meningitis, chagas disease and respiratory syncytial virus infection in infected infants, children below 5 years and adults [11, 15]. However, findings from case–control studies in malaria endemic countries regarding association of these polymorphisms with susceptibility to clinical and severe malaria have been contradictory [16–23]. These discrepancies have been based on the differences geographical location and genetic background of human populations where these studies were conducted [16, 23]. Therefore, there is a need for more immunogenetic studies, regarding the role of TLR4 polymorphisms in malaria pathogenesis, particularly in other countries with high malaria transmission.
Nigeria is currently among the top there high malaria burden countries in the world with an annual mortality of 300,000 deaths and where falciparum malaria accounts for 30 % of total under-five mortality every year [24]. Despite evidence from the HapMap project that the various genotypes of TLR4 Asp299Gly and Thr399Ile SNPs are in circulation among the Yoruba tribe [25], the roles of these SNPs in influencing susceptibility to clinical and severe malaria remain unknown. It is on this basis that the present study was carried out to determine the frequency, distribution and association of mutant genotypes of TLR4 Asp299Gly and Thr399Ile polymorphisms in a cohort of P. falciparum infected Nigerian children with susceptibility to clinical and severe malaria.
Methods
Study design and settings
This was a cross sectional study in which convenience sampling was used to enroll children during two separate malariometric surveys in Lagos between 2009 – 2011, the first survey was conducted in March – August, 2009 in Takway-Bay, Victoria Island, Lagos, while the second survey was conducted during the dry season in January 2011 in Ibeshe Community in Ikorodu. To further obtain TLR4 SNPs data from severe malaria cases, children hospitalized at Massey Street Children Hospital, Lagos with laboratory and clinical evidence of severe falciparum malaria between September – October, 2011 were also enrolled into the study. Lagos is located within the Equatorial tropical region where malaria transmission mostly driven by female mosquitoes from the Anopheles gambie complex occurs throughout the year [26]. In Ibeshe, malaria prevalence rate of 14.2 % (95 % CI, 13.3 – 16 %) has been previously reported by Aina et al. [27], while in Takwa-Bay, children below 10 years account for 76.2 % of cases of uncomplicated malaria cases seen (Iwalokun et al., unpublished). In this coastal settlement, malaria treatment practices were generally poor among caregivers despite their good knowledge of symptoms of uncomplicated malaria [28]. Massey Street Children Hospital is a foremost referral health facility of children in south West Nigeria where management of severe malaria on yearly basis is very common [29].
Study population
The study population for this study comprised children (age < 13y) with Plasmodium falciparum positivity slide results. Asymptomatic malaria was defined as parasitaemia without fever in the previous 48 h or other related malaria symptoms or history of malaria in the preceding two months. Children in the uncomplicated malaria category were those having parasitaemia with fever (axillary temperature > 37.4 °C) other classical symptoms such as headache, chill and sweating plus mild-moderate anaemia (Hb < 11 g/dL - <9 g/dL), while severe malaria category were those with severe anaemia, fever plus one or more of other complications such as prostration, coma, jaundice, and respiratory distress [30]. At the time of enrollment, anthropometric measurements were performed with the children wearing light clothing and no shoes. Body length of children up to 23 months old was measured in recumbent position using a wooden horizontal stadiometer, while height of children aged 24 months and above was measured using a vertical stadiometer. These children were weighed using appropriate balance to the nearest 0.1 kg. The anthropometric measurements were then transformed into z scores with the aid of Epi-info 2000 software version 3.4 [31] and used for comparison with growth curve published by National Centre for Health statistics (NCHS) [32]. The z-score values for height for age (HAZ), weight for age (WAZ) and weight for height (WHZ) < −2 SD were defined as stunting, under weight and wasting based on the NCHS indices [32]. The field enrolled children with asymptomatic and uncomplicated malaria were treated with arthemether-Lumefantrine according to the national treatment guidelines, while those with severe malaria were referred to Massey Street Children’s Hospital for care with intramuscular loading dose of arthemether (3.2 mg/kg) given as a pre-referral treatment [33]. Children who did not have Yoruba ethnic background, with parasite negative glass slides and those whose caregivers declined consent were excluded from the study. This study was ancillary to the malariometric study that was approved by the institutional review board of the Nigerian Institute of Medical Research (NIMR-IRB), Lagos –Nigeria.
Parasite detection
Parasite was detected and speciated by microscopic examination of thick (12 μL) and thin (3 μL) blood smear on grease-free labeled glass slides according to WHO guidelines. The glass slides were prepared in duplicates and read by two independent trained microscopists. A glass slide was considered parasite negative after examining 200 high power fields without seeing one parasite. Parasitaemia was measured by counting the number of parasites against 200–500 leukocytes using the thick blood film and expressed as number of parasites per 1 microlitre of whole blood, assuming 8000 leukocytes per 1 microlitre of blood.
Blood haemoglobin measurement
A drop of finger pricked blood drawn into a microcuvette was used for the determination of haemoglobin (g/dL) using a Hemocue machine (Hemocue Hb 201). Anaemia was defines as Hb below 11 g/dL and in this study, Hb <7 g/dL was taken as severe malaria.
Genomic DNA extraction
The salting out protocol of Miller et al. [34] was used for the extraction and purification of genomic DNA from peripheral blood samples of the study participants collected separately into labeled EDTA vials. DNA purification was done using the phenol-chloroform method.
TLR4 (Asp299Gly and Thr399Ile) genotyping
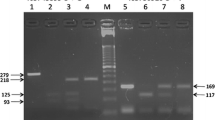
The alleles and genotypes of the two studied TLR4 SNPs were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) based method using the previously used primers sequences by Nyati et al. [35] (Table 1). All PCR amplification reactions were performed in a 20-uL volume PCR tubes containing 10X PCR buffer, 200 μM each of the dNTPs, 2.0 mM of MgCl2, 20 pmol of each primer, 1.25 U of Taq DNA polymerase (Promega, USA) and 100 ng of each genomic DNA as template. In a thermal cyler (Techne™ Thermal cycler TC-312, Fisher Scientific, UK), the PCR reaction was then subjected to denaturation at 94 °C for 5 min, followed by 35 cycles of denaturing at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min and cooling to 4 °C. Template-free water was used as a negative control. After amplification, the purified 4-μL PCR products were subjected to restriction digestion by NcoI restriction endonuclease (Fermentas) for TLR4 Asp299Gly and by HinfI for TLR4 Thr399Ile with 1 mL 10X enzyme buffer (Fermentas) overnight at 37 °C. The PCR products yielded 299 bp and 406 bp respectively, while the digested products yielded same DNA fragments for the wild alleles but decreased to 233 bp and 377 bp fragments for the 299Gly and 399Ile mutant alleles respectively, on 3 % agarose gel after electrophoresis (Table 1). Each restriction enzyme assay was duplicated for the confirmation of RFLP results.
Statistical analysis
Data were double-entered into Microsoft Excel 2008 and Microsoft Access 2008 and validated before analysis using SPSS statistical software for windows version 15.0 (SPSS Inc, USA). Data were expressed as number and percentages (%), median and range, mean ± standard error of mean (SEM) and 95 % confidence intervals (95 % CI). High parasite density and hyperparasitaemia were defined as parasitaemia > 10,000/uL and > 250,000/uL respectively. Axillary body temperature > 40 °C was indicative of hyperpyrexia, while severe malnutrition was defined as WHZ or WAZ or HAZ < −3 relative to NCHS references. Disparity in mean values between asymptomatic control and clinical malaria case was measured by Student’s t-test, while allele and genotype frequency comparison was evaluated by chi-square (χ2) test using SPSS version 15.0 for windows. Association of TLR4 Asp299Gly and Thr399Ile polymorphisms with clinical and severe malaria was measured by calculating odd ratio (OR) and their 95 % confidence intervals (95 % CI) using dominant inheritance model [23]. This was due to very low numbers of homozygous mutant genotypes of both TLR4 Asp299Gly and Thr399Ile polymorphisms. Therefore, they were grouped together with their respective heterozygotes as mutant genotypes for the calculation of odd ratios and their 95 % CI between clinical malaria cases and the asymptomatic control and between severe and uncomplicated malaria. Odd ratios and their 95 % CI without and with adjustment were also used to measure association of the TLR4 SNPs with the malaria disease phenotypes (i.e. anaemia, severe anaemia, malnutrition, severe malnutrition, high parasite density severe) measured as covariates. Adjustment was performed by including only cases of comparable ages between ASM and clinical malaria for anaemia, malnutrition (WHZ or WAZ or HAZ < −2) and gender or age for severe anaemia, severe malnutrition and high parasite density seen only in clinical malaria cases for the analysis of genotype distribution of the TLR4 SNPs. Hardy-Weinberg Equilibrium (HWE) was conducted by comparing of the observed frequencies of different genotypes of the two TLR4 SNPs with their expected frequencies under HWE for each study group and outcomes with P > 0.05 was considered to be in HWE [36]. Genotypic deviation of HWE was measured by Pearson’s chi-square (χ2) statistical test. All statistical outcomes with P-value < 0.05 were considered to be significant.
Results
A total of 279 children comprising 182 children (62.1 % male; mean ± SEM age, 57.3 ± 1.7 months) with clinical falciparum malaria and 97 children (55.7 % male; mean ± SEM age, 55.6 ± 2.5 months) with asymptomatic falciparum malaria were enrolled (Table 2).
The prevalence rates of underweight, stunting and wasting were found to be 22.3 %. 23.8 % and 3.7 % among the study participants. Among the children with clinical malaria, 2 (1.1 %), 5 (2.7 %), 15 (8.2 %) elicited hyperparasitaemia, hyperpyrexia and severe malaria. Between asymptomatic and clinical malarial children there was no statistical significance difference (P < 0.05) in terms of age, sex and being underweight. But variables such as stunting, wasting and axillary body temperature and anaemia occurred more significantly (P <0.05) in symptomatic children (Table 2).
All the genotypes of both TLR4 SNPs were found in the study population (Tables 3 & 4). However, the difference in the proportions of genotypes of these SNPs between clinical malaria and asymptomatic control cases was not significant (P > 0.05) (Table 3). Further stratification of the clinical malaria cases showed that the frequencies of the minor alleles: 299Gly and 399Ile, of these TLR4 SNPs were greater than 10 % (17.6 % and 14.7 % respectively) only in severe malaria children (Table 3 & 4).
Unlike in asymptomatic population, the genotype distribution of TLR4 Asp299Gly polymorphism was not in HWE in the clinical malaria group but did not condition susceptibility (Tables 3 & 4).
However, with stratification, Asp299Gly and Thr399Ile polymorphisms were found to increase the risk of severe malaria 3-fold and 8-fold respectively (P < 0.05) (Table 4). Further analysis revealed mutant genotypes of these SNPs to also increase the risk of severe anaemia, high parasite density and severe malnutrition 3.8 -5.3-fold, 3.3 – 4.4-fold and 4-fold respectively (Table 5).
Discussion
The present study has examined the role of the two non-synonymous SNPs, rs4986790 (Asp299Gly) and rs4986791 (Thr399Ile), located at the third exon of the TLR4 gene in the development of clinical and severe malaria among children with Yoruba ethnic background. This selection was based on evidence of existence of the two SNPs of TLR4 among the Yoruba population (http://www.hapmap.org) [25]. For the first time in the setting of P. falciparum malaria, this study has also found all the genotypes of TLR4 SNPs among children of Yoruba ethnicity. However, the difference in the numbers of mutant genotypes of both TLR4 Asp299Gly and Thr399Ile SNPs between asymptomatic and clinical malarial children was observed to be non-significant (P < 0.05), translating to lack of association between carriage of these genetic variants and clinical malaria in the studied children. Our finding is similar to the report of Esposito et al. [37]. The workers also did not find significant association between the carriage of TLR4 Asp299Gly SNP and malaria among Burundian children. The MAF of 6 % for 299Gly reported for the uninfected and malarial children by these workers is also similar to 5.4 – 7 % range found in this study. However, unlike in the Burundian children, we found MAF of 17.6 % and 10.4 % for TLR4 Asp299Gly and Thr399Ile among our severe malaria sub-group with genotypes containing these alleles further found to increase the risk of severe malaria 3-fold and 8-fold respectively. Our findings indicate that the two minor alleles 299 Gly and 399Ile of TLR4 SNPs are not protective against severe malaria in the studied children. This is in agreement with the case–control studies conducted in Ghana [21]. In the Ghanian study, Mockhenhaupt et al. [21] reported also frequencies of 17.6 % and 24.1 % for these minor alleles in severe malarials children compared to 2.4 % and 6.2 % in healthy control. The workers also found these minor alleles to confer 1.5- and 2.6-fold increased risk of severe malaria respectively. In this study, we found the Asp299Gly heterozygote genotype in 8.7 %, 9.4 % and 23.5 % among the studied children with asymptomatic, UM and SM. Meanwhile, contrary to our findings and those of Mockenhaupt et al. [21], neither TLR4 Asp299Gly nor Thr399Ile polymorphism was found to be associated with the risk of severe malaria anaemia and cerebral malaria in children from Uganda and Cameroun [17, 23]. This is in spite of the location of these countries in sub-Saharan Africa. This may not be unconnected with difference in ethnic background of these children and sample size coupled with temporal changes in malaria transmission, occurring in many malaria endemic African countries that might explain the different genotype and allele frequencies of TLR4 Asp299Gly and Thr399Ile polymorphisms reported by these investigators. For instance, in the Cameroun study of 1,862 children, Apinjo et al. [23] reported MAF of 0.2 – 8.1 % for both TLR4 polymorphisms in asymptomatic and clinical malaria (uncomplicated and severe malaria) children from Bantu, Foulbe and Semi-Bantu ethnic groups. The Ugandan study was conducted among 137 P. falciparum infected hospitalized children and only the frequency of Asp299Gly was above 10 % (i.e. 12.3 %) in the cerebral malaria group that also had a frequency of 1.5 % for the Thr399Ile genotype. This genotype was not found in children with uncomplicated malaria. In Brazil, where temporal changes from low to moderate P. vivax and P. falciparum malaria transmission also occur, contradictory findings, regarding the association of TLR4 Asp299Gly polymorphisms with protection from clinical malaria have been reported [19, 20]. Within the Amazon region, da Silva et al. [19] found MAF of 5.8 % and 1.5 % for TLR4Asp299Gly polymorphism among 113 healthy and 535 clinical malaria participants with this polymorphism eliciting protection against clinical malaria, while Soares et al. [20] did not find protective effect of TLR4Asp299Gly among their 44 study participants in an earlier study.
Taken together, our findings and previous reports on TLR4 Asp299Gly and Thr399Ile polymorphisms in malaria endemic countries strongly point to the relevance of other infectious diseases that also provide selection pressure on TLR4. They include tuberculosis, HIV, gram negative bacterial infection, candidiasis, meningitis and respiratory syncytial viral infection [5, 11]. Since the burden of these infectious diseases varies across the various malaria endemic countries, there is thus a high possibility of differences in their contributions to the evolution of mutant variants of TLR4.
Therefore, the implications of other TLR4 activated infectious diseases stated above highlights one of the limitations of the present study since the children were enrolled during malariometric surveys and were not clinically and diagnostically examined for candidemia and viral infections that may further influence the phenotypic disposition of TLR4 polymorphism and subsequently impact malaria susceptibility.
In this study and under dorminant model of inheritance, the significant association of the two TLR4 SNPs with clinical malaria syndromes such as fever, severe anaemia and high parasite density was found. This findings indicate that both TLR SNPs elicit functional effects that are related to the pathogenesis of falciparum malaria in the studied children. Similar phenotypic findings were also reported by Mochenhaupt et al. [22] in Ghanian pregnant women, the population of whom are also at high risk of clinical malaria globally. The workers found TLR4Asp299Gly polymorphism to increase maternal anemia 4.7-fold after adjustment by age. Here, we have found both TLR4 Asp299Gly and Thr399Ile polymorphisms to cause a 3.8 -5.3-fold increased risk of severe anaemia and 3.5 – 4.4-fold increased risk of high parasite density, while Thr399Ile polymorphism alone raised the risk of fever 5-fold. In addition to Mockhenhaupt et al. [21], The phenotypic effects of TLR4 polymorphisms found in these study suggest that functional variations may exist between Asp299Gly and Thr399Ile SNPs of TLR4, regarding their systemic effects in Nigerian children with clinical malaria. Future studies that will look the combined effects of these SNPs through haplotype analysis will be very important. On the contrary, the functional effects of the two TLR4 SNPs observed in this study and in previous Ghanian study were absent in Camerounian children [23]. This disparity again confirm that malaria is a complex disease, involving multiple genetic factors that play different biological roles in disease manisfestation. Therefore, future haplotype-malaria association studies are needed to resolve inconsistency or heterogenous genotype-phenotype relationships in the setting of malaria in the African region.
However, of relevance is our finding that the mutant genotypes of TLR4Asp299Gly increased the risk of severe malnutrition 3.7-fold. However, this observation needs to be interpreted with caution because it was not been documented in previous immunogenetic studies of TLR4 and malaria from other countries. Also because of the fact that malnutrition is a common health problem among African children, including Nigeria and many aetiologies have been reported [38, 39]. In fact, it has been reported that malnutrition accounts for 22 – 40 % of under –five mortality and malaria is one of the co-morbid factors [40, 41]. In the study, 3.7 %, of our study population as a whole elicited wasting, which indicates acute malnutrition 23.8 % were stunted, indicating chronic malnutrition and a state of long period of nutrient deprivation, while 22.3 % were underweight. Even in the setting of P. falciparum infection, these rates appear to be lower than the national prevalence rates 5.5 %, 35.7 % and 25.2 % and rates reported for children below 5 years in the northern parts of Nigeria. Another reason for this cautionary interpretation is the fact the present study is cross-sectional in design and is limited in determining the cause and effect relationship between malnutrition and malaria in the studied children. furthermore, the malnutrition investigated in this study exluded micronutrient deficiency such as zinc, iron and vitamin A deficiency, which have been shown to increase the risk of severe anaemia, wasting and dyregulated innate immunity in malarial children [42].
In conclusion, our findings suggest that TLR4 Asp299Gly and Thr399Ile polymorphisms among Nigerian children of Yoruba ethnic background may modulate susceptibility to severe malaria and other disease outcomes such as severe anaemia and hyperpyrexia. To further validate the present findings, functional studies of TLR4 polymorphisms, haplotype analysis and investigation of other TLR4 activated infectious diseases in the setting of malaria in this category of Nigerian children are needed.
References
World Health Organisation (2012). World Malaria Report 2012. Available: http://www.who.int/malaria/publications/world_malaria_report_2012/report/en/index.html. Accessed 2013 June08.
Douglas J, Perkins, Tom W, Gregory C, Davenport, Hittner JB, et al. Severe malarial anemia: innate immunity and pathogenesis. Int J Biol Sci. 2011;7(9):1427–42.
Malaria GEN. A global network for investigating the genomic epidemiology of malaria. Nature. 2008;456:732–8.
Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4:169–80.
Ziakas PD, Prodromou ML, El Khoury J, Zintzaras E, Mylonakis E. The role of TLR4 896 A > G and 1196 C > T in susceptibility to infections: a review and meta-analysis of genetic association studies. PLoS One. 2013;8, e81047.
Othoro C, Lal AA, Nahlen B, Koech D, Orago AS, Udhayakumar V. A low interleukin-10 tumor necrosis factor-alpha ratio is associated with malaria anemia in children residing in a holoendemic malaria region in western Kenya. J Infect Dis. 1999;179:279–82.
Achidi EA, Apinjoh TO, Yafi CN, Besingi R, Anchang JK, Awah NW, et al. Plasma Levels of Tumour Necrosis Factor-Alpha, Interleukin-10, Interleukin-12, Macrophage Inhibition Factor and Transforming Growth Factor-Beta in Children with Severe and Uncomplicated Falciparum Malaria. J Trop Dis. 2013;1:103.
Perkins DJ, Were T, Davenport GC, Kempaiah P, Hittner JB, Ong'echa JM. Severe malarial anaemia: innate immunity and pathogenesis. Int J Biol Sci. 2011;7:1427–42.
Hughes S, Kelly P. Interactions of malnutrition and immune impairment with specific reference to immunity against parasites. Parasite Immunol. 2006;28:577–88.
Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76.
Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci U S A. 2007;104:16645–50.
Krishnegowda G, Hajjar AM, Zhu J, Douglass EJ, Uematsu S, Akira S, et al. Induction of proinflammatory responses in macrophages by the Glycosylphosphatidylinositols of Plasmodium falciparum: cell signaling receptors, Glycosylphosphatidylinositol (GPI) structural requirement, and regulation of GPI activity. J Biol Chem. 2005;280:8606–16.
Prohinar P, Rallabhandi P, Weiss JP, Gioannini TL. Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J Immunol. 2010;184:4362–7.
Coban C, Ishii KJ, Uematsu S, Arisue N, Sato S, Yamamoto M, et al. Pathological role of Toll-like receptor signaling in cerebral malaria. Int Immunol. 2007;19:67–79.
Löfgren J, Marttila R, Renko M, Rämet M, Hallman M. Toll-like receptor 4 Asp299Gly polymorphism in respiratory syncytial virus epidemics. Pediatr Pulmonol. 2010;45:687–92.
Basu M, Maji AK, Chakraborty A, Banerjee R, Mullick S, Saha P, et al. Genetic association of Toll-like-receptor 4 and tumor necrosis factor-alpha polymorphisms with Plasmodium falciparum blood infection levels. Infect Genet Evol. 2010;10:686–96.
Sam-Agudu NA, Greene JA, Opoka RO, Kazura JW, Boivin MJ, Zimmerman PA, et al. TLR9 polymorphisms are associated with altered IFN-gamma levels in children with cerebral malaria. Am J Trop Med Hyg. 2010;82:548–55.
Zakeri S, Pirahmadi S, Mehrizi AA, Djadid ND. Genetic variation of TLR-4, TLR-9 and TIRAP genes in Iranian malaria patients. Malar J. 2011;10:77.
da Silva Santos S, Clark TG, Campino S, Suarez-Mutis MC, Rockett KA, Kwiatkowski DP, et al. Investigation of host candidate malaria-associated risk/protective SNPs in a Brazilian Amazonian population. PLoS One. 2012;7(5):e36692.
Soares SC, Abé-Sandes K, Nascimento Filho VB, Nunes FM, Silva Jr WA. Genetic polymorphisms in TLR4, CR1 and Duffy genes are not associated with malaria resistance in patients from Baixo Amazonas region, Brazil. Genet Mol Res. 2008;7(7):1011–9.
Mockenhaupt FP, Cramer JP, Hamann L, Stegemann MS, Eckert J, Oh NR, et al. Toll-like receptor (TLR) polymorphisms in African children: common TLR-4 variants predispose to severe malaria. J Commun Dis. 2006;38:230–45.
Mockenhaupt FP, Hamann L, von Gaertner C, Bedu-Addo G, von Kleinsorgen C, Schumann RR, et al. Common polymorphisms of toll-like receptors 4 and 9 are associated with the clinical manifestation of malaria during pregnancy. J Infect Dis. 2006;194:184–8.
Apinjoh TO, Anchang-Kimbi JK, Njua-Yafi C, Mugri RN, Ngwai AN, Rockett KA, et al. Achidi EA; MalariaGEN Consortium Association of cytokine and Toll-like receptor gene polymorphisms with severe malaria in three regions of Cameroon. PLoS One. 2013;8, e81071.
National Population Commission (NPC) (Nigeria), National Malaria Control Programme (NMCP) (Nigeria), ICF International. Nigeria Malaria Indicator Survey 2010. Abuja, Nigeria: NPC, NMCP, and ICF International. 2012.
International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320.
Awolola TS. Okwa, Hunt RH, Ogunrinade AF, Coetzee M. Dynamics of the malaria-vector populations in coastal Lagos, south-western Nigeria. Ann Trop Med Parasitol. 2002;96:75–82.
Aina OO, Agomo CO, Olukosi YA, Okoh HI, Iwalokun BA, Egbuna KN, et al. Malariometric Survey of Ibeshe Community in Ikorodu, Lagos State: Dry Season, Malaria Research and Treatment. 2013; 13: Article ID 487250. http://dx.doi.org/10.1155/2013/487250Research.
Iwalokun BA, Agomo PU, Egbuna KN, Iwalokun SO, Adebodun V, Olukosi OO, et al. Environmental Survey and Health Seeking Behavior of Caregivers of Children Suspected to have Malaria inTakwa-Bay, Lagos State. J Med Sci. 2011;2:675–87.
Afolabi BM, Clement CO, Ekundayo A, Dolapo D. A hospital-based estimate of major causes of death among under-five children from a health facility in Lagos, Southwest Nigeria: possible indicators of health inequality. Int J Equity Health. 2012;11:39.
WHO. Severe falciparum malaria. World Health Organization, Communicable Diseases Cluster. Trans R Soc Trop Med Hyg. 2000;94(1):S1–90.
National Guidelines for Diagnosis and Treatment of Malaria. Federal Ministry of Health, National Malaria and Vector Control Division, Abuja, Nigeria; 2011. Available from:http://www.nmcpnigeria.org/f/casemanagement/National%20Guidelines%20on%20Diagnosis%20%20&Treatment%20of%20Malaria%20in%20Nigeria%20June%202011.pdf. Accessed February 12, 2012
Dean AG, Dean JA, Burton AH, Dicker RC. Epi Info™: A general purpose microcomputer program for health information systems. Am J Preventive Med. 1991;7:178–82.
National Center for Health Statistics (NCHS). Growth Curves for Children Birth: 18 years. United States: Department of Health, Education and Welfare, Publication 78, 1. 1997.
Miller SA, Dykes DD, Polesky HF. A Simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1998;16:1215.
Nyati KK, Prasad KN, Verma A, Singh AK, Rizwan A, Sinha S, et al. Association of TLR4 Asp299Gly and Thr399Ile polymorphisms with Guillain-Barre’ syndrome in Northern Indian population. J Neuroimmunol. 2010;218:116–9.
Michael C: A simple calculator to determine whether observed genotype frequencies are consistent with Hardy-Weinberg equilibrium. 2008 http://www.tufts.edu/~mcourt01/Documents/Courtlab- HW calculator.xls
Esposito S, Molteni CG, Zampiero A, Baggi E, Lavizzari A, Semino M, et al. Role of polymorphisms of toll-like receptor (TLR) 4, TLR9, toll-interleukin 1 receptor domain containing adaptor protein (TIRAP) and FCGR2A genes in malaria susceptibility and severity in Burundian children. Malar J. 2012;11:196.
Olumese PE, Sodeinde O, Ademowo OG, Walker O. Protein energy malnutrition and cerebral malaria in Nigerian children. J Trop Pediatr. 1997;43:217–9.
Tine RC, Ndiaye M, Hansson HH, Ndour CT, Faye B, Alifrangis M, et al. The association between malaria parasitaemia, erythrocyte polymorphisms, malnutrition and anaemia in children less than 10 years in Senegal: a case control study. BMC Res Notes. 2012;5:565.
Ibekwe VE, Ashworth A. Management of protein energy malnutrition in Nigeria: an evaluation of the regimen at the Kersey Nutrition Rehabilitation Center, Nigeria. Trans R Soc Trop Med Hyg. 1994;88:594–5.
Ubesie AC, Ibeziako NS, Ndiokwelu CI, Uzoka CM, Nwafor CA. Under-five protein energy malnutrition admitted at the University of Nigeria Teaching Hospital, Enugu: a 10 year retrospective review. Nutr J. 2012;11:43.
Ling PR, Schwartz JH, Bistrian BR. Mechanisms of host wasting induced by administration of cytokines in rats. American J Physiology. 1997;272:E333–9.
Acknowledgement
We thank the patients and their caregivers for their co-operation as well as the field health workers for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that there are no conflicts of interest regarding this work.
Authors’ contributions
BAI conceptualized the study idea, carried out molecular and haematological assays and drafted the manuscript. AO was involved in parasite detection, site preparation, manuscript preparation and supervision of data management. SOI was involved in patients’ enrollment, stratification, and treatment, anthropometric measurement and manuscript preparation. PA was also involved in study design. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Iwalokun, B.A., Oluwadun, A., Iwalokun, S.O. et al. Toll-like receptor (TLR4) Asp299Gly and Thr399Ile polymorphisms in relation to clinical falciparum malaria among Nigerian children: a multisite cross-sectional immunogenetic study in Lagos. Genes and Environ 37, 3 (2015). https://doi.org/10.1186/s41021-015-0002-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41021-015-0002-z