Abstract
Background
Glioma is the most common malignant brain tumor in adults. The standard treatment scheme of glioma is surgical resection combined alternative radio- and chemotherapy. However, the outcome of glioma patients was unsatisfied. Here, we aimed to explore the molecular and biological function characteristics of GPX7 in glioma.
Methods
The multidimensional data of glioma samples were downloaded from Chinese Glioma Genome Atlas (CGGA). RT-qPCR method was used to identify the expression status of GPX7. Kaplan–Meier curves and Cox regression analysis were used to explore the prognostic value of GPX7. Gene Set Enrichment Analysis (GSEA) was applied to investigate the GPX7-related functions in glioma.
Results
The results indicated that the expression of GPX7 in glioma was higher compared to that in normal brain tissue. Univariate and multivariate Cox regression analyses confirmed that the expression value of GPX7 was an independent prognostic factor in glioma. The GSEA analysis showed that GPX7 was significantly enriched in the cell cycle pathway, ECM pathway, focal adhesion pathway, and toll-like receptor pathway.
Conclusions
The GPX7 was recommended as an independent risk factor for patients diagnosed with glioma for the first time and GPX7 could be potentially used as the therapy target in future. Furthermore, we attempted to explore a potential biomarker for improving the diagnosis and prognosis of patients with glioma.
Similar content being viewed by others
Background
Glioma is a malignant tumor with the highest morbidity and mortality in the central nervous system, which represented up to 81% of central nervous system malignancies [1, 2]. The current standard treatment is surgical resection, combined radiotherapy and chemotherapy [3]. Although active treatments are taken, the overall survival (OS) and progression-free survival of glioma patients is still unsatisfying and unpredictable [4].. Therefore, it is imperative to identify the novel prognostic biomarkers for glioma.
Considerable studies have demonstrated that mRNA is involved in the molecular regulation mechanism of glioma. Thus, mRNA can be used as a potential biomarker to predict the overall survival time of glioma patients [5]. For instance, LGALS3, L1CAM, and SCAMP3 were associated with the shorten OS of glioma patients by promoting the proliferation or other malignant tumor characteristics of glioma [6,7,8]. Despite previous studies partially revealed that the abnormal expression of mRNA is implicated in the occurrence and development of glioma via multiple mechanisms, the pathogenesis of these mRNAs remains limited due to the complex biological characteristics in gliomas. Therefore, the aim of this study was to explore a crucial mRNA for glioma to predict the prognosis of glioma patients.
Mammalian GPX7 is a non-selenocysteine containing phospholipid peroxide glutathione peroxidase [9] and plays a significant role in maintaining redox status [10]. Therefore, the body avoids entering an oxidative stress state. Numerous studies have revealed that oxidative stress state exhibited an inhibitory effect on the pathological process of tumors. For example, miRNA-338-5p promotes oxidative stress by regulating the Hedgehog pathway and ultimately inhibited the proliferation of gliomas [11]. Moreover, inducing oxidative stress of C6 glioma cells was related with the promotion of cell cycle arrest and apoptosis [12]. However, whether GPX7 was involved in the pathological process of tumors remains unknown. Peng et al. showed that GPX7 was absent in esophageal adenocarcinoma and was associated with methylation of specific sites in the exon region of GPX7. Besides, the biological effect of overexpressing GPX7 on inhibiting cell proliferation and promoting cell senescence has been verified in vitro experiments and in vivo models [13]. However, the expression of GPX7 was also reported as an oncogene. Nonetheless, recent studies have showed that overexpression of GPX7 could predict malignant entities in liver cancer [14]. Hence, the role of GPX7 in tumors remains controversial. So far, there is no relevant report of the function of GPX7 in glioma. Thus, the role of GPX7 in glioma, whether it could promote the pathological process of glioma or serves as a clinical therapeutic target, remained elusive.
Here, we investigated the molecular and clinical features of GPX7 in glioma through bioinformatics analysis. Meanwhile, we attempted to reveal the GPX7-related pathological progress in gliomas. At the same time, we endeavored to establish a novel therapeutic target to improve the diagnosis and the prognosis of glioma.
Methods
RNA sequencing and clinic information from CGGA data repository
We downloaded the clinical and multidimensional data of more than 1000 Chinese glioma patients from the Chinese Glioma Genome Atlas (CGGA) (http://www.cgga.org.cn/). The data included the whole-exome sequencing (286), DNA methylation (159), mRNA sequencing (1018), mRNA microarray (301), microRNA microarray (198), and matched clinical data. In CGGA database, the RNA sequencing data and clinical information of 325 and 693 patients were obtained in two sessions. The sva package and limma package in R language were used to intersect the genes of the two data sets and merge the obtained data into one data set for subsequent analysis. Further, the glioma samples with incomplete clinical information were eliminated. And finally, 749 patients were enrolled in our study.
Gene expression profiling interactive analysis (GEPIA) database (http://gepia.cancer-pku.cn/) is a web server for gene expression profiling and interaction analysis of cancer and normal tissues. A total of 163 samples of glioblastoma, 518 samples of low-grade glioma, and 207 normal samples were enrolled in our study.
GSEA
Gene Set Enrichment Analysis (GSEA) is a useful tool which could be used to analyze the functions and signal pathways of genes. The samples downloaded from the CGGA database were separated into high GPX7-expression group and low GPX7-expression group. A p value of less than 0.05 and an FDR value of less than 0.25 were significant.
RT-qPCR
Total RNA was extracted from gliomas and normal control tissues by tri-Regent (Sigma, USA) (samples source: the First Affiliated Hospital of Harbin Medical University), RT-qPCR was performed using fast start universal SYBR Green Master (Rox) (Roche, Germany), -Δ CT reckoned the mRNA expression. The sequence of GAPDH was 5′-CAAGGTCATCACTGATGAACTTTG-3′ (F) and 5′-GTCCACCCTGTTGCTGTAG-3′ (R), and the primer sequence of GPX7 was 5′-TCACAGACCACTACCGA-3′ (F) and 5′-CGGGGACACTACTCATTC-3′ (R).
Statistical analysis
The transcriptome data from glioma samples from the CGGA database were comprehensively analyzed. Kaplan–Meier method and Cox regression model were used to exploring the prognostic value of GPX7 in glioma patients. Gene set enrichment analysis (GSEA) was used to predict the GPX7-related biological functions and pathways in gliomas. Co-expression analysis was used to detect the genes most related to the expression of GPX7.
Results
Clinical characteristics
The clinical information of gliomas, including PRS type, histology, grade, gender, age, radio status, chemotherapy, IDH mutation status, and 1p19q codeletion status, were shown in Table S1.
High expression of GPX7 shortens the overall survival time in glioma patients
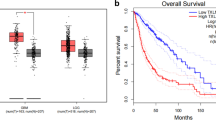
To explore how GPX7 affects glioma patients, we separated the obtained samples into high GPX7 expression group and a low GPX7 expression group. The survival package and survminer package were used to analyze the survival differences between two group. The five-year survival rate of the GPX7 overexpression group was 14.7% (95% CI [0.1107–0.196]) while the 5-year survival rate of the GPX7 low expression group was 63.3% (95% CI [0.583–0.689]) (Fig. 1a).
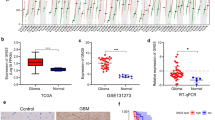
The Kaplan–Meier survival curve and The ROC curve. a Highly expressed GPX7 leads to shorter overall survival time for patients with glioma. b ROC curve shows that using GPX7 as a prognostic marker is valuable. c The expression of GPX7 in GBM and LGG was higher than that in normal tissues in the GEPIA database. d Glioma patients with high GPX7 expression have shorter survival times than those with low expression in the GEPIA database
The receiver operating characteristic curve (ROC) was used to further verify whether the expression level of GPX7 was reliable in predicting the survival time of patients. The AUC was more than 0.7 in the 1-year, 3-year, and 5-year ROC curve for the prognostic model (Fig. 1b). The area under the ROC curve (AUC) was more significant than 0.7, and which suggested that the GPX7 expression was reliable in predicting survival time.
Additionally, according to the GEPIA database, the expression of GPX7 was significantly higher in glioblastoma (GBM) and low-grade gliomas (LGG) than normal tissues. What is more, high GPX7 expression remains associated with poor prognosis of glioma patients (p < 0.05) (Fig. 1c, d).
Overexpressed GPX7 as an independent risk factor for predicting the prognosis of glioma patients
Furthermore, to verify whether GPX7 can be used as an independent risk factor in predicting the prognosis of glioma patients, the Cox regression model was used to assess GPX7 expression and other clinical features. Univariate and multivariate Cox analysis confirmed that GPX7 was an independent risk factor for glioma patients. The remaining clinical features associated with the OS of glioma patients are shown in Fig. 2.
The relationship between GPX7 and different clinical characteristics
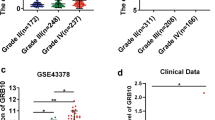
Based on previous studies, the factors that might influence the prognosis of glioma patients were identified. Then, we used the Wilcox.test and Kruskal.test to investigate the relationship between GPX7 and clinical factors. The results revealed that the expression of GPX7 was positively correlated with PRS type, history, grade, age, and chemotherapy, while it was negatively correlated with IDH mutation status and 1p19q codeletion status (Fig. 3a-g).
Potential pathway of GPX7 in regulating malignant biological behavior of glioma
We used the GSEA method to identify the GPX7-related biological functions. Notably, GSEA is a ubiquitous bioinformatics analysis tool. This can help researchers identify the enrichment pathway of the target gene and indirectly understand the biological function of the target gene. Therefore, GPX7 was divided into a high expression group and a low expression group. The results indicated that the GPX7 expression was related to multiple tumor-related pathways, including the cell cycle pathway, ECM pathway, focal adhesion pathway, and toll-like receptor pathway (Fig. 4a–d). These pathways were thus considered as potential strategies for GPX7 to achieve its regulation in glioma.
Co-expression analysis
To further clarify the role of GPX7 in the pathophysiology of gliomas, we found the GPX7-related genes with the Pearson correlation test. Top five genes which were positively correlated with GPX7, including DDOST, RCC2, MCM3, MAGOH, and HDAC1, were selected. Meanwhile, the top five genes which were negatively correlated with GPX7, including RIMS1, AMER3, TMEM56, SVOP, and FBXW4 were selected. As shown in Figs. 5 and 6, the result suggested that GPX7 might promote the expression of genes that are positively correlated with its expression level or that these co-expressed positively related genes co-promote or inhibit specific pathophysiological processes with GPX7 to promote the occurrence and development of glioma. Conversely, GPX7 might inhibit the expression of genes that are negatively correlated, or the function between them is antagonistic.
Validating GPX7 expression level in glioma using RT-qPCR
By analyzing the data from the CGGA database, it was concluded that GPX7 is a novel oncogene, and its overexpression indicated a poor prognosis for gliomas. For further verification, 5 glioma samples and 5 normal brain tissue samples were used to verify the expression level of GPX7 by RT-qPCR. As a result, the expression of GPX7 in gliomas was higher compared to that in normal tissues (p < 0.05) (Fig. 7), which corroborated with the findings from previous analysis of the CGGA database.
Discussion
Reports indicate that GPX7 functions distinctly in different tumors. For instance, it is a tumor suppressor gene in esophageal adenocarcinoma but a carcinogenic gene in liver cancer [13, 14]. Nonetheless, the role of GPX7 in glioma remains unknown. As such, to reveal the role of GPX7 in glioma, this work first obtained RNA sequencing data of more than one thousand glioma patients from the CGGA database.
Through the correlation analysis between GPX7 and OS of glioma patients, we discovered that the is the expression of GPX7 was negatively related with the OS in glioma patients. It is of pivotal importance to verify the reliability of GPX7 in predicting the OS of glioma patients by the ROC curve. The overexpression of GPX7 in gliomas was further validated by the GEPIA database and it was associated with poor prognosis. Nevertheless, the occurrence and development of glioma are induced by the imbalance between multiple carcinogenic and cancer suppressive factors; hence, several factors are implicated in this complex pathological process. Whether GPX7 can be used as an independent risk factor for glioma patients remains unclear.
Univariate and multivariate analyses were performed by the Cox regression model. As a result, we found that GPX7 could still be used as an independent risk factor in predicting the prognosis of glioma patients after excluding the influence of other factors. At the same time, our analysis demonstrated that other clinical characters, including PRS type, grade, age, chemotherapy, IDH mutation status, and 1p19q codeletion status, can also act as independent risk factors. Moreover, we uncovered that the expression level of GPX7 is related to different clinical characteristics, thereby indicating that overexpression of GPX7 might influence the malignant progress of these clinical characteristics via some potential mechanisms. Based on the above studies, we proved for the first time that GPX7 could be used as a novel oncogene in predicting the prognosis of glioma patients. Nonetheless, the mechanism of GPX7 in regulating the malignant behavior of glioma remains controversial.
GSEA analysis suggested that GPX7 might involve in joining the pathophysiological process of glioma. The outcomes revealed some significantly enriched pathways, including cell cycle pathway, ECM pathway, focal adhesion pathway, and toll-like receptor pathway, Catchin flavonoids inhibited glioma by blocking cell cycle signaling pathways [15]. The invasiveness of glioma cells can be achieved by selectively interacting with the extracellular matrix (ECM) components [16]. Mda-9/syntenin promotes glioma migration through focal adhesion kinase (FAK) in the focal adhesion signaling pathway [17]. The toll-like receptor family is closely related to cancer. Toll-like receptor 4 promotes glioma proliferation by activating the NF-kappa B signaling pathway [18]. Toll-like receptor 2 regulates glioma invasion by promoting the expression of matrix metalloproteinases [19]. The above pathways promote the pathological process of glioma through proliferation, migration, invasion, and other malignant biological behaviors of glioma cells. The enrichment of GPX7 in these pathways revealed that glioma might participate in the regulation via various complex pathways. Despite reaching a preliminary conclusion on how GPX7 regulates glioma by GSEA analysis, the biological behavior of glioma cannot be fully regulated by a single factor due to the complex pathological process of glioma.
Therefore, we screened 10 genes having the highest correlation with GPX7 expression by co-expression analysis, among which 5 genes had a positive correlation, including DDOST, RCC2, MCM3, MAGOH, and HDAC1. The pathophysiological processes regulated by these genes were closely related to tumors. The high expression of DDOST in colon adenocarcinoma promotes the proliferation of tumor cells [20]. The expression of RCC2 in ER-positive breast induces the expression of IGF-1 and leads to the malignant behavior of tumor cells, including proliferation, invasion, etc. [21]. MCM3 is an independent risk factor for invasive ductal carcinoma [22]. MAGOH regulates melanoma production by controlling the proliferation of melanocyte [23]. HDAC1 promotes liver cancer metastasis via the FAM99A-miR92a signaling pathway [24]. Additionally, the 5 negatively related genes include RIMS1, AMER3, TMEM56, SVOP, and FBXW4. Therefore, the occurrence and development of glioma is a significantly complex biological process, and GPX7 interacts with various oncogenes and tumor suppressor genes to participate in the network of glioma regulation. GPX7 is considered as a novel molecular marker that predicts the prognosis of glioma patients, with high validity and reliability.
For the first time, through analysis of multidimensional data, we propose that GPX7 can be used as a novel oncogene to shorten the prognosis of glioma patients. Nonetheless, this study has some shortcomings. Firstly, the CGGA database only enrolled more than 1000 high-throughput data of glioma patients without non-glioma individuals. Therefore, all the findings were based on the comparison between the high- and low-expression of the GPX7 group. However, we found that the high expression of GPX7 accounts for malignant progression of glioma by comparing the overall survival time between the two groups and verifying its credibility via ROC curves. The expression of GPX7 was validated in glioma and normal brain tissues by RT-qPCR to compensate for the insufficiency of sample information in the database. Consequently, we found that GPX7 was highly expressed in glioma. Notably, this was in line with our previous findings.
Conclusion
For the first time, the GPX7 was reported as a carcinogenic gene in glioma, and our results revealed the correlation between gliomas and malignant clinical features. Moreover, our findings provided a new diagnostic and prognostic marker for study in future.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- CGGA:
-
The Chinese Glioma Genome Atlas
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- OS:
-
Overall survival
- GSEA:
-
Gene Set Enrichment Analysis
- ROC:
-
Receiver operating characteristic curve
- AUC:
-
Area under the ROC curve
References
Zhang C, Cheng W, Ren X, et al. Tumor Purity as an Underlying Key Factor in Glioma. Clin Cancer Res. 2017;23(20):6279–91.
Adel Fahmideh M, Schwartzbaum J, Frumento P, et al. Association between DNA repair gene polymorphisms and risk of glioma: a systematic review and meta-analysis. Neuro Oncol. 2014;16(6):807–14.
Cheng M, Zhang ZW, JI XH, et al. Super-enhancers: A new frontier for glioma treatment. Biochim Biophys Acta Rev cancer. 2020;1873(2):188353.
Chen W, Lei C, Liu P, et al. Progress and Prospects of Recurrent Glioma: A Recent Scientometric Analysis of the Web of Science in 2019. World Neurosurg. 2020;134:e387–e99.
Öberg K, Califano A, Strosberg JR, et al. A meta-analysis of the accuracy of a neuroendocrine tumor mRNA genomic biomarker (NETest) in blood. Ann Oncol. 2020;31(2):202–12.
Hu WM, Yang YZ, Zhang TZ, et al. LGALS3 Is a Poor Prognostic Factor in Diffusely Infiltrating Gliomas and Is Closely Correlated With CD163+ Tumor-Associated Macrophages. Front Med. 2020;7:182.
Zeng J, Xi SY, Wang F, et al. L1CAM High Expression Associates with Poor Prognosis in Glioma but Does Not Correlate with C11orf95-RELA Fusion. BioMed Res Int. 2020;2020:1353284.
Li C, Zhang Z, LV P, et al. SCAMP3 Promotes Glioma Proliferation and Indicates Unfavorable Prognosis via Multiple Pathways. Onco Targets Ther. 2020;13:3677–87.
Utomo A, Jiang X, Furuta S, et al. Identification of a novel putative non-selenocysteine containing phospholipid hydroperoxide glutathione peroxidase (NPGPx) essential for alleviating oxidative stress generated from polyunsaturated fatty acids in breast cancer cells. J Biol Chem. 2004;279(42):43522–9.
Chang YC, Yu YH, Shew JY, et al. Deficiency of NPGPx, an oxidative stress sensor, leads to obesity in mice and human. EMBO Mol Med. 2013;5(8):1165–79.
Zhou H, Han L, Wang H, et al. Chidamide Inhibits Glioma Cells by Increasing Oxidative Stress via the miRNA-338-5p Regulation of Hedgehog Signaling. Oxid Med Cell Longev. 2020;2020:7126976.
Shukla A, Trivedi SP. An in vitro analysis of the rat C6 glioma cells to elucidate the linear alkylbenzene sulfonate induced oxidative stress and consequent G2/M phase cell cycle arrest and cellular apoptosis. Chemosphere. 2018;205:443–51.
Peng D, Hu T, Soutto M, et al. Glutathione peroxidase 7 has potential tumour suppressor functions that are silenced by location-specific methylation in oesophageal adenocarcinoma. Gut. 2014;63(4):540–51.
Guerriero E, Capone F, Accardo M, et al. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur J Histochem. 2015;59(4):2540.
Zhan Sheng Z. Anticancer effects of catechin flavonoid in human glioma cells are mediated via autophagy induction, cell cycle arrest, inhibition of cell migration and invasion and targeting MAPK/ERK signalling pathway. J BUON. 2020;25(2):1084–90.
Goldbrunner RH, Bernstein JJ, Tonn JC. ECM-mediated glioma cell invasion. Microsc Res Tech. 1998;43(3):250–7.
Zhong D, Ran JH, Tang WY, et al. Mda-9/syntenin promotes human brain glioma migration through focal adhesion kinase (FAK)-JNK and FAK-AKT signaling. Asian Pac J Cancer Prev. 2012;13(6):2897–901.
Jiang Y, Zhou J, Luo P, et al. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NF-κB signaling pathway. EBioMedicine. 2018;37:78–90.
Wang F, Zhang P, Yang L, et al. Activation of toll-like receptor 2 promotes invasion by upregulating MMPs in glioma stem cells. Am J Transl Res. 2015;7(3):607–15.
Hu M, Fu X, Si Z, et al. Identification of Differently Expressed Genes Associated With Prognosis and Growth in Colon Adenocarcinoma Based on Integrated Bioinformatics Analysis. Front Genet. 2019;10:1245.
Wang W, Xu B, Zhang Z, et al. RCC2 Expression Stimulates ER-Positive Breast Tumorigenesis. J Oncol. 2020;2020:5619462.
Zhao Y, Wang Y, Zhu F, et al. Gene expression profiling revealed MCM3 to be a better marker than Ki67 in prognosis of invasive ductal breast carcinoma patients. Clin Exp Med. 2020;20(2):249–59.
Silver DL, Leeds KE, Hwang HW, et al. The EJC component Magoh regulates proliferation and expansion of neural crest-derived melanocytes. Dev Biol. 2013;375(2):172–81.
Zhao B, Ke k, Wang Y, et al. HIF-1α and HDAC1 mediated regulation of FAM99A-miR92a signaling contributes to hypoxia induced HCC metastasis. Signal Transduct Target Ther. 2020;5(1):118.
Acknowledgements
Not applicable.
Funding
National Natural Science Foundation of China (81972363).
Author information
Authors and Affiliations
Contributions
JW Y and XC designed and guided the subject. ZD L and RT Z collated and analyzed the data. CZ and QY completed the writing and repair of the manuscript. PL Y and QY J completed the validation experiment part. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Informed consent was obtained from patients to collect tissue samples for the study, and the study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University (No. HMUIRB-2008-06).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1: Table S1.
Characteristics of patients with glioma based on CGGA. Description of data: Clinical information of the samples we used for analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yao, J., Chen, X., Liu, Z. et al. The increasing expression of GPX7 related to the malignant clinical features leading to poor prognosis of glioma patients. Chin Neurosurg Jl 7, 21 (2021). https://doi.org/10.1186/s41016-021-00235-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41016-021-00235-3