Abstract
Commonly observed aberrations in epidermal growth factor receptor (EGFR) signaling have led to the development of EGFR-targeted therapies for various cancers, including non–small cell lung cancer (NSCLC). EGFR mutations and overexpression have further been shown to modulate sensitivity to these EGFR-targeted therapies in NSCLC and several other types of cancers. However, it is clear that mutations and/or genetic variations in EGFR alone cannot explain all of the variability in the responses of patients with NSCLC to EGFR-targeted therapies. For instance, in addition to EGFR genotype, genetic variations in other members of the signaling pathway downstream of EGFR or variations in parallel receptor tyrosine kinase (RTK) pathways are now recognized to have a significant impact on the efficacy of certain EGFR-targeted therapies. In this review, we highlight the mutations and genetic variations in such genes downstream of EGFR and in parallel RTK pathways. Specifically, the directional effects of these pharmacogenetic factors are discussed with a focus on two commonly prescribed EGFR inhibitors: cetuximab and erlotinib. The results of this comprehensive review can be used to optimize the treatment of NSCLC with EGFR inhibitors. Furthermore, they may provide the rationale for the design of subsequent combination therapies that involve the inhibition of EGFR.
Similar content being viewed by others
Introduction
Non–small cell lung cancer (NSCLC) accounts for nearly 80% of all lung cancers and is the leading cause of cancer-related deaths worldwide [1,2]. Moreover, late-stage detection limits the treatment options for many patients with NSCLC given that most cancers have already metastasized at the time of diagnosis [1]. In more than one half of all patients with NSCLC, the aberrant epidermal growth factor receptor (EGFR) signaling contributes to the oncogenic phenotype [1]. More recent attempts to treat NSCLC have thus focused on targeting EGFR in order to abrogate the oncogenic signaling mediated by activating EGFR mutations (found in approximately 15% of patients with NSCLC), EGFR overexpression, and/or EGFR gene copy number enhancement [3-5]. For example, EGFR inhibition is achieved through two main classes of drugs: tyrosine-kinase inhibitors (TKIs) and monoclonal antibodies. Cetuximab (Erbitux™) is a commonly prescribed monoclonal antibody for the treatment of metastatic NSCLC. Cetuximab inhibits EGFR by binding to its extracellular domain, which then blocks ligand-dependent receptor activation [6]. Although less clearly understood, cetuximab also inhibits EGFR signaling by mediating receptor endocytosis and degradation and thus it also decreases ligand-independent EGFR signaling [7]. On the contrary, erlotinib (Tarceva™) is a frequently prescribed TKI for the treatment of NSCLC. By binding to the intracellular kinase domain of EGFR at the ATP-binding site, erlotinib inhibits kinase activity by blocking ATP hydrolysis [1,8-10].
Pharmacogenomic studies have shown that EGFR mutation status is associated with erlotinib efficacy and that EGFR overexpression is associated with patient response to cetuximab and other EGFR-targeted agents [1,11-13]. However, even among patients who are selected for specific treatments based on their somatic EGFR mutation status or EGFR expression profile, there remains a notable lack of response to EGFR-targeted therapies in a significant portion of the patient population. For instance, approximately 30% of patients with NSCLC with activating EGFR mutations do not respond as expected to TKIs against EGFR [1,14]. Therefore, although EGFR status is still an important indicator of patient response to EGFR-targeted therapies, it is clearly not the only gene that influences the therapeutic response. A review of the pharmacogenomics of cetuximab and erlotinib instead reveals that other genetic factors, beyond EGFR, influence the efficacy of these agents and can potentially guide the treatment of NSCLC with mutant EGFR or EGFR overexpression. In fact, cetuximab serves as a model candidate drug with which to explore the effects of such non-EGFR genetic variations on the treatment of NSCLC given the established association between Kirsten rat sarcoma viral oncogene (KRAS) mutations and poor efficacy of cetuximab in the treatment of colorectal cancer [15]. Similar non-EGFR genetic variations have been implicated in modulating erlotinib efficacy in those patients with NSCLC who harbor activating EGFR mutations [1,16,17]. More specifically, recent and compelling evidence now suggests that genetic variations in other members of the signaling pathway downstream of EGFR, and also in the non-EGFR receptor tyrosine kinase (RTK) pathways, can influence responses to cetuximab and erlotinib.
The EGFR signaling network
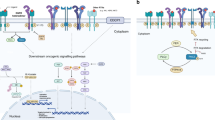
EGFR signaling contributes to the regulation of fundamental biological processes including cell proliferation, differentiation, survival, adhesion, homeostasis, and tumorigenesis [18-21]. Exceedingly complex and highly regulated signal transduction mechanisms are required to govern such varied EGFR responses to external stimuli [19,20,22]. Given the vast complexity of the EGFR signaling network, it is hardly surprising that genetic factors beyond EGFR mutations or variable expression patterns may modulate therapeutic responses to EGFR-targeted agents. Here, we present an overview of EGFR signaling and highlight the primary downstream signaling pathways (Figure 1).
Schematic representation of the primary epidermal growth factor receptor (EGFR) signaling pathway. During normal EGFR signaling, receptor activation is dependent on ligand-mediated receptor dimerization. Once the subunits dimerize, a series of phosphorylation events serve to enhance EGFR kinase activity to stimulate the activation of downstream targets. Downstream signals are propagated by EGFR through three central pathways via both the direct phosphorylation of downstream targets (the JAK/STAT pathway) and the membrane recruitment of key adaptor proteins (the PI3K/AKT and MAPK pathways) to promote cell survival and proliferation. The EGFR signaling through a conserved core of three downstream signaling pathways demonstrates how the activation of this pathway via parallel RTKs, such as HER2, HER3, and MET, can circumvent the inhibitory effects of cetuximab and erlotinib on EGFR. EGFR, epidermal growth factor receptor; JAK/STAT, Janus activated kinase/signal transducer and activator of transcription; PI3K/AKT, phosphoinositide 3-kinase/protein kinase B; MAPK, mitogen-activated protein kinase; HER2, human epidermal growth factor receptor 2; HER3, human epidermal growth factor receptor 3; MET, hepatocyte growth factor receptor; SOS, son of sevenless; GRB2, growth factor receptor-bound protein 2; RAS, rat sarcoma family of proteins; RAF, rapidly accelerated fibrosarcoma; MEK, MAPK kinase; PTEN, phosphatase and tensin homolog; mTOR, mammalian target of rapamycin; FOXO, forkhead box proteins; NF-κB, nuclear factor-kappa B.
EGFR is a membrane-spanning cell surface receptor and consequently links internal signaling pathways to the extracellular environment [19]. When stimulated by extracellular ligand binding, EGFR can initiate intracellular kinase cascades through its intracellular tyrosine kinase domain (Figure 1) [18,19]. In humans, 7 peptide growth factors function as EGFR ligands, among which the canonical ligand is epidermal growth factor (EGF) [18,21]. These ligands can either be membrane-bound to adjacent cells or soluble if they undergo proteolysis at the cell membrane, which allows EGFR to integrate stimuli from a variety of local and distant sources (Figure 1) [21,23].
Structurally, EGFR has a domain architecture that consists of four extracellular domains, one transmembrane domain, one juxtamembrane domain, and one tyrosine kinase domain followed by a flexible C-terminal tail (Figure 1) [24]. Prior to ligand binding, EGFR exists as a catalytically inactive monomer in the cell membrane [19,24]. Following bivalent ligand binding to a single EGFR molecule, the receptor changes conformation to facilitate dimerization and activation [18,19,24]. Receptor activation then proceeds through a defined series of auto and transphosphorylation events between the kinase domains in the EGFR dimer [19]. The activated kinases further proceed to phosphorylate the C-terminal tail regions on the neighboring receptor, creating phosphorylated tyrosine (pY) residues [19,25]. The pY residues then serve as docking sites for proteins that are capable of binding pY motifs (Figure 1) [19,20,26]. The docked proteins can then either transduce intracellular signals following direct phosphorylation by EGFR or initiate higher-order signaling cascades through scaffolding and localization mechanisms (Figure 1) [19,27,28].
Once activated, EGFR transduces its numerous cellular responses through three primary signaling cascades: the mitogen–activated protein kinase (MAPK) pathway, the phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) pathway, and the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway (Figure 1) [1,18,25]. Notably, these three central signaling pathways, although downstream of EGFR, are also activated by almost every other RTK (Figure 1) [19,29]. Once EGFR activates the rat sarcoma (RAS) family of proteins, the MAPK pathway proceeds through the consecutive phosphorylation of rapidly accelerated fibrosarcoma (RAF), MAPK kinase (MEK), and MAPK via the recruitment of the son of sevenless/growth factor receptor-bound protein 2 (SOS/GRB2) complex (Figure 1) [18,30]. The MAPK cascade results in the activation of various transcription factors, which act to alter cellular expression profiles and to promote proliferation (Figure 1) [30]. The PI3K/AKT pathway not only governs the anti-apoptotic and pro-survival signals associated with EGFR activation but also plays a role in the regulation of cell growth via the activation of the mammalian target of rapamycin (mTOR) (Figure 1) [1,31]. Moreover, AKT activation promotes the accumulation of nuclear factor-kappa B (NF-κB) in the nucleus, which further promotes cell survival [32]. The PI3K/AKT pathway is also negatively regulated by phosphatase and tensin homolog (PTEN), a phosphatase that opposes PI3K activity to repress AKT activation (Figure 1) [33]. JAK/STAT signaling is initiated when proteins of the JAK family are recruited to EGFR and are activated via phosphorylation [34]. Activated JAK proteins can phosphorylate proteins of the STAT family, which then dimerize, translocate to the nucleus, and enact global cellular effects, particularly those related to cell survival and proliferation (Figure 1) [34]. Clearly, the downstream effects of EGFR signaling reveal the oncogenic potential of aberrant EGFR signaling.
Review
Erlotinib and cetuximab as EGFR-targeted therapeutics in NSCLC: pharmacogenomics of EGFR
The genetic variation in EGFR and how this variation alters typical EGFR signaling, plus its effects on cetuximab and erlotinib efficacy, have all been extensively reviewed elsewhere [1,35,36]. However, it is important to briefly describe the genetic variations in EGFR in order to provide a framework for the interpretation of how non-EGFR genetic variations can affect responses to cetuximab and erlotinib; namely, aberrant activation of signaling pathways downstream of, or parallel to, the EGFR signaling network can compensate for EGFR inhibition. On the most basic level, two types of genetic variations in EGFR drive hyperactive signaling: constitutive activation and overexpression [1,36]. EGFR molecules can be constitutively active due to either point mutations in the kinase domain or larger deletions in the juxtamembrane domain that relieve the autoinhibition of the ligand-free monomer [1,19,36,37]. Thus, even in the absence of ligand, these receptors are active and continually promote downstream signaling. EGFR overexpression is another mechanism that contributes to hyperactive EGFR signaling. The observed increase in signaling strength from EGFR overexpression is partly due to the increase in the number of receptors, which amplify the downstream signaling effects [1,36,38]. In addition, the overexpression of wild-type (wt)-EGFR can promote rogue signaling through a ligand-free activation mechanism comparable to that of mutant EGFR. Specifically, the increased local concentration of EGFR at the cell membrane can increase the stochastic, ligand-free dimerization of EGFR that activates the receptors [39].
Unlike newer TKIs that irreversibly bind to EGFR, erlotinib reversibly binds to the ATP-binding pocket of EGFR and inhibits catalytic activity via the prevention of ATP-dependent activation of downstream targets [8,40]. Moreover, erlotinib is very similar to gefitinib, another commonly studied EGFR TKI, in that they have similar mechanisms of action and have been demonstrated to have similar effectiveness in the treatment of NSCLC, despite reports of minor differences in toxicity [41,42]. Given its mechanisms of action and the prerequisite of ATP hydrolysis for EGFR catalytic activity, erlotinib was predicted to elicit a positive drug response in patients with NSCLC as long as they were positive for EGFR expression, regardless of the EGFR mutation status. However, a wealth of research has been conducted that suggests that only certain activating mutations in EGFR serve as positive clinical indicators for the use of erlotinib in patients with NSCLC [25,43-46]. Surprisingly, it has been demonstrated that while several mutations in EGFR confer a susceptibility to erlotinib, others facilitate the opposite effect—a resistance to erlotinib [47-49]. The most common activating mutations in EGFR, which account for 90% of all EGFR mutations in cases of NSCLC, are exon 19 deletions and the L858R mutation in the tyrosine kinase domain (Table 1) [1]. Other point mutations such as G719C, G719S, G719A, and L861Q similarly result in mutant forms of EGFR that are hyperactive (Table 1) [1]. Importantly, all of these activating mutations have been shown to incur sensitivity to EGFR TKIs such as erlotinib (Table 1) [1]. Other activating EGFR mutations, primarily in exon 20, such as T790M (gatekeeper mutation), L747S, D761Y, and T854A, also result in overactive EGFR signaling but instead grant resistance to erlotinib and other TKIs that are similar to erlotinib [1,48,50,51]. More importantly, given their association with acquired TKI resistance, these mutations appear to overrule the normal erlotinib sensitivity incurred by the more common EGFR-activating mutations when both mutation types are concurrently present (Table 1) [1,48]. As a consequence, several irreversible potent anti-EGFR agents have either been approved by the United States Food and Drug Administration (FDA) (e.g., afatinib) or show promise in clinical trials to overcome the T790M-mediated EGFR TKI resistance [52-55]. The variability in the response to erlotinib in the treatment of NSCLC is further accentuated by the fact that only approximately 10% of patients without activating EGFR mutations demonstrate a drug response to first-line erlotinib treatment [1,56-58]. Clearly, the gene-drug relationship of EGFR and erlotinib is dependent upon more than EGFR mutation status alone, and these findings suggest the need to elucidate the predictive value of other specific, non-EGFR genetic variants that could guide the treatment of NSCLC.
Some studies suggest that the response to erlotinib is also improved in patients with EGFR overexpression [1,59]. However, this association might be due to the link between the efficacy of erlotinib and EGFR mutation status because activating EGFR mutations often promote increased expression and altered copy number of EGFR. Moreover, other studies have provided contrary evidence with regard to the association between EGFR expression and benefits from erlotinib treatment [1]. Unlike erlotinib, cetuximab has an efficacy that has been associated with high levels of EGFR (mutant or wild-type) expression but not with EGFR mutations or copy number variation (Table 1) [11,13]. Variation in EGFR expression alone, though, is not sufficient to explain the pharmacogenomics of cetuximab in the treatment of NSCLC. For example, one trial evaluated the expression of immunohistochemically detectable EGFR as a biomarker, and recruited patients to examine the effectiveness of cetuximab in combination with first-line chemotherapy [60,61]. The trial showed that standard chemotherapy plus cetuximab was superior to chemotherapy alone in patients whose tumors expressed EGFR [60,61]. In contrast to these findings, the analysis of a similar trial that lacked patient selection based on EGFR expression status found no association of EGFR expression status with cetuximab benefit; however, this study confirmed that EGFR mutation status and gene copy number are not predictive biomarkers of the response to cetuximab treatment [60-63]. Despite these inconsistencies, a meta-analysis of 4 such trials has provided evidence that a high level of EGFR expression can predict clinical benefits from cetuximab therapy [13]. Therefore, unlike EGFR mutation status for erlotinib, the most appropriate biomarker for the selection of patients with NSCLC for cetuximab treatment appears to be the expression level of EGFR. However, further studies are still needed to corroborate these findings given the debate over the cutoff between high and low expression and because several studies still question the predictive value of the expression level of EGFR in the treatment of patients with NSCLC [6,64,65].
Pharmacogenomics beyond EGFR: downstream pathway members
Clear pharmacogenomic associations exist between cetuximab and erlotinib and genes other than EGFR. Some of the most well-characterized associations involve genetic variations in downstream pathway members in the EGFR signaling network. Given that the signaling pathways that are activated by EGFR primarily consist of kinase phosphorylation cascades, it is not surprising that mutations in these kinases are similar to those that result in the constitutive activation of EGFR. As shown in Figure 1, EGFR regulates numerous signaling pathways after activation and thus mutations in any downstream pathway member could foreseeably compensate for EGFR inhibition via the overstimulation of a specific node of downstream signaling.
Prime examples of genetic variation in signaling proteins downstream of EGFR are KRAS mutations (Table 1) [1,16]. KRAS mutations have been linked to the reduced efficacy of erlotinib in the treatment of NSCLC and to that of cetuximab in the treatment of colorectal cancer [1,15,17]. Moreover, evidence from other RTK-targeted TKIs suggests that mutations in KRAS are highly associated with intrinsic resistance to TKI treatment [1,17]. For instance, one study found that 95% of patients with NSCLC who had detectable KRAS mutations were resistant to the treatment with TKIs and showed continued disease progression [1]. In addition, multiple studies have demonstrated that tumors with activating KRAS exon 1 mutations at G12X and G13X are associated with a lack of response to erlotinib and gefitinib (Table 1) [17,66]. Notably, resistance to erlotinib does not appear to be modulated by specific subtypes of activating KRAS mutations, unlike the variable erlotinib sensitivities incurred by different EGFR mutations [67,68]. Granted, while most of the evidence supports that KRAS mutations grant resistance to EGFR-targeted TKIs, some debate remains concerning the prognostic value of the KRAS genotype on TKI treatment of NSCLC. This is especially true given that in rare cases, patients with KRAS mutations still respond to erlotinib [66,69]. Although KRAS mutations are generally thought of as mutually exclusive to EGFR mutations, overexpression of (wt)-EGFR appears to be associated with a response to erlotinib in these patients with KRAS mutants. This could explain the atypical drug sensitivity exhibited by these patients [1,66,69].
With respect to cetuximab for the treatment of colorectal cancer, the negative prognostic value of KRAS mutations is rigorously defined and is similar to the lack of a response to erlotinib in KRAS-mutant patients with NSCLC [15]. However, current evidence suggests that KRAS mutations in patients with NSCLC do not necessarily predict a poor response to cetuximab treatment [16,68]. Instead of contradicting the importance of KRAS variability in the treatment of NSCLC, the different prognostic values of KRAS mutations in colorectal cancer versus NSCLC only confirm the need to determine how non-EGFR genetic variations affect the outcome of cetuximab treatment. For example, one convincing explanation for the variable responses to cetuximab in patients with KRAS-mutant colorectal cancer compared with patients with KRAS-mutant NSCLC is that the KRAS mutations found in each cohort are different and could thus elicit variable protein-level effects on the structure and function of KRAS. More specifically, a significantly larger number of KRAS-activating G > T DNA transversions were found at codon 12 of the KRAS gene in patients with NSCLC compared with those found in patients with colorectal cancer [68,70]. This might be due to the association of such DNA transversion events with tobacco-related carcinogens [16,68,70]. The KRAS transversion mutations in patients with NSCLC correspond to a greater frequency of the G12C variant compared with the more predominant G12D variant found in patients with colorectal cancer [68]. Interestingly, recent findings have shown that in the setting of colorectal cancer, at least 12 subtypes of KRAS mutations across codons 12 and 13 associate with variable yet negative efficacy of cetuximab [68,71]. An understanding of what the different KRAS mutations are in NSCLC and how they modulate the efficacy of cetuximab could perhaps reveal similar subsets of KRAS mutations with specific predictive value to treatment options of NSCLC.
If KRAS mutations are the most predominant genetic alterations in the MAPK pathway that influence the efficacy of EGFR-targeted therapies, then variation in the phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) is perhaps the corresponding genetic abnormality in the downstream portion of the PI3K/AKT pathway (Table 1) [1]. PIK3CA encodes the p110α subunit of PI3K and is thus an integral part of the PI3K/AKT signaling pathway (Figure 1) [1,31]. Previous evidence has shown that PIK3CA mutations frequently co-exist with EGFR mutations in NSCLC and that these mutations could be predictive of reduced sensitivity to TKIs (Table 1) [1,72-74]. Two of the most frequent mutations in the helical domain of PIK3CA, E542K and E545K, as well as one mutation in the kinase domain, H1047R, are known to activate the p110α subunit and to stimulate oncogenic signaling via the PI3K/AKT pathway (Table 1) [73,75]. Notably, when such activating PIK3CA mutations are introduced into EGFR-mutant lung cancer cells, they impart a partial resistance to TKIs (Table 1) [73-76]. Interestingly, while this in vitro association between PI3KC mutations and enhanced resistance to EGFR-targeted TKIs has yet to be replicated in the clinic in patients with NSCLC, one recent study confirmed the high frequency of co-mutations of PI3KC and EGFR in patients with NSCLC [72]. The same study also found that the status of PI3KC mutations was a negative prognostic indicator for survival in patients with (wt)-EGFR and (wt)-KRAS subtypes [72]. Moreover, although limited experimental data are available to explore the exact prognostic value of PIK3CA mutations on the efficacy of cetuximab in NSCLC, activating PIK3CA mutations in only the p110α catalytic domain, and not in the helical domain, appear to grant similar resistance to cetuximab in colorectal cancer cohorts (Table 1) [77]. Moreover, other studies involving human head and neck cancer cell lines found that treatment with either a PI3K inhibitor or a dual PI3K/mTOR inhibitor enhances the sensitivity of cancer cells to cetuximab in vitro and in vivo [78,79]. Thus, PI3K mutations may contribute to the reduced efficacy of erlotinib and cetuximab in specific subtypes of cancer cells; however, further investigation is needed to confirm these associations in the clinical setting of NSCLC.
Given the important regulatory role of PTEN in AKT activation (Figure 1), it is not surprising that PTEN is another gene from the PI3K/AKT pathway with genetic variations that are associated with the efficacy of EGFR-targeted drugs [74]. Namely, one screen of various NSCLC cell lines revealed that the homozygous loss of PTEN was associated with the resistance to erlotinib in the H1650 cell line [80]. In agreement with that report, another study found that PTEN overexpression was associated with prolonged survival after TKI treatment in patients with NSCLC, which suggests that low expression or deletion of PTEN is associated with shortened survival in patients with NSCLC [81]. PTEN also plays a role in the modulation of the response to cetuximab. For instance, NSCLC cell lines were found to acquire resistance to cetuximab, as well as to erlotinib, as a result of enhanced PTEN instability and degradation [82]. This finding is notable in that it highlights how, apart from the presence of a clear homozygous PTEN deletion, PTEN genotyping may be less effective than PTEN expression testing for the determination of actual PTEN levels. Additional studies of the effects of PTEN expression on the response to cetuximab in patients with metastatic colorectal cancer corroborate the results in NSCLC in that the loss of PTEN expression was found to be associated with poor overall survival and drug resistance during cetuximab therapy [83,84].
Genetic variations in the NF-κB transcription factor (Figure 1), a downstream member of the PI3K/AKT pathway, also associate with specific responses to erlotinib and cetuximab (Table 1) [1]. Specifically, the nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (NFKBIA) gene encodes the inhibitor of NF-κB (IκBα) protein, which inhibits NF-κB activity by binding NF-κB and confining it to the cytoplasm [1,85,86]. Strong evidence supports that the knockdown of NFKBIA confers partial resistance to erlotinib in lung cancer cell lines through elevated levels of NF-κB and that this resistance can be reversed by the inhibition of NF-κB or by the overexpression of NFKBIA (Table 1) [1,87]. It was also demonstrated that NFKBIA expression is a successful prognostic biomarker for patients with EGFR-mutant NSCLC who are treated with erlotinib and that low NFKBIA expression is predictive of poor progression-free survival and overall survival [1,87]. Notably, NFKBIA silencing is also found far more frequently in patients with NSCLC who lack EGFR mutations than in those with EGFR-mutant NSCLC, which suggests that such downstream activation of the PI3K/AKT pathway can bypass the need for mutations in upstream driver proteins during carcinogenesis [85]. However, the mechanism that surrounds the variable expression of NFKBIA has not been extensively studied. Further support for the importance of NF-κB comes from studies on astrocyte elevated gene-1 (AEG-1), an oncogene that can increase NF-κB activity by activating the IκB kinase (IKK), a protein that functions to destabilize IκBα (Table 1) [1]. Patients with NSCLC whose tumors express higher levels of AEG-1 appear to have poorer outcomes than those with lower levels of AEG-1 after treatment with TKIs, which suggests that genetic variations in AEG-1 could alter EGFR-targeted drug response in a similar fashion to NFKBIA (Table 1) [1]. With respect to cetuximab, while no specific evidence links genetic variations in NF-κB or NFKBIA with drug efficacy on NSCLC, patients with colorectal cancer whose tumors are negative for NF-κB expression have a positive predictive response to cetuximab and a longer overall survival compared with patients whose tumors are positive for NF-κB expression; the results were similar after erlotinib treatment in patients with NSCLC (Table 1) [1,88].
Pharmacogenomics beyond EGFR: Non-EGFR RTKs
Just as genetic variations in the members of the EGFR signaling pathway can bypass EGFR inhibition when downstream effectors of EGFR are activated, mutations in, or the overexpression of, non-EGFR RTKs can also thwart the efficacy of EGFR-targeted agents via the activation of parallel signaling pathways. While the term “parallel” is used to signify that a different RTK initiates the signaling cascade, it is important to emphasize that almost all RTKs function to activate the same small number of central pathways (Figure 1) [29]. Therefore, downstream of RTK activation, these parallel pathways merge with the previously described pathways downstream of EGFR (Figure 1). Significantly, there are only 58 known RTKs among the 90 tyrosine kinases in the human genome [19,89]. Given the redundancy of the pathways downstream of activation, aberrant activation of any other RTKs could foreseeably hinder a patient’s response to erlotinib or cetuximab. This can therefore provide a mechanism for both primary and acquired resistance to these EGFR-targeted agents.
EGFR-targeted therapies in NSCLC have been linked to genetic abnormalities in the MET gene, which encodes the MET protein or hepatocyte growth factor receptor (HGFR), an RTK that recognizes the hepatocyte growth factor (HGF) ligand [90]. In lung cancer, various mutations in both the juxtamembrane domain and the extracellular domains of MET, along with MET amplification, are known to activate the receptor and stimulate the PI3K/AKT signaling [91]. For instance, one study found that increased MET copy number and MET overexpression are negative prognostic factors for surgically resected NSCLC (Table 1) [1,92]. In addition, MET gene amplification and overexpression were found to be associated with resistance of NSCLC cell lines to both erlotinib and gefitinib (Table 1) [93]. MET activation via HGF ligand stimulation has also been shown to induce resistance to EGFR-targeted TKIs even in the presence of activating EGFR mutations (Table 1) [94]. Notably, gene amplification of MET has been identified in up to 20% of EGFR-mutant tumors that have been pretreated with EGFR-targeted TKIs (Table 1). This suggests a role for MET in the mediation of acquired resistance to these drugs since MET amplification was rarely concomitant with the common resistance-granting T790M EGFR mutation [1,76,91,95,96]. Furthermore, one study detailed a mechanism for such MET-mediated resistance by showing that an EGFR TKI-sensitive lung cancer cell line can develop resistance to gefitinib as a result of MET amplification via human epidermal growth factor receptor 3 (HER3)-dependent activation of PI3K (Table 1) [97]. Similarly, while the associations between cetuximab and MET are currently limited to the setting of colorectal cancer, MET amplification and activation were associated with acquired resistance to cetuximab in treated patients, and cetuximab-induced MET activation was found to contribute to cetuximab resistance in certain colon cancer cell lines (Table 1) [98,99].
Beyond its role in the modulation of overactive MET signaling, HER3, a kinase-impaired RTK in the EGFR family, can form active heterodimers with other members of the EGFR family, namely EGFR and human epidermal growth factor receptor 2 (HER2) [1,100]. It thus stands to reason that HER3 expression patterns and mutation status could impact the efficacy of therapies based on EGFR inhibition. Indeed, one study showed elevated levels of not only MET but also HER2 and HER3 in cells that developed an acquired resistance to cetuximab [101]. The authors propose that cetuximab treatment induces the up-regulation of EGFR, which allows EGFR to form heterodimers with HER3, HER2, and MET. This allows for the maintenance of downstream EGFR signaling despite the inhibition of EGFR homodimers [101]. The same study further supports such a heterodimer-mediated mechanism of resistance by demonstrating strong anti-proliferative effects when these cancer cells were treated with both an inhibitor of HER2/3 heterodimerization and cetuximab, which suggests that HER2/3 heterodimers also help mediate cetuximab resistance in an EGFR-dependent manner (Table 1) [101]. Given that HER2 is the preferred dimerization partner of HER3 and that HER2/3 heterodimers are more active and have greater oncogenic potential than other dimers of the EGFR family, it is logical that HER2/3 dimers form and drive rogue signaling once both HER2 and HER3 are recruited to the membrane by EGFR [102]. Therefore, such findings detail one method, similar to those described for MET-mediated resistance, by which cells can evade EGFR inhibition from cetuximab. This is primarily accomplished by the re-establishment of downstream tumorigenic signaling via coordination with alternative RTKs.
The trend of up-regulated RTK activity that modulates resistance to EGFR-targeted therapies is also observed with HER2. For example, enhanced copy number of the HER2 gene is associated with increased resistance to both erlotinib and cetuximab (Table 1) [103,104]. Moreover, activating HER2 mutations are similarly associated with poor response to erlotinib, even when they are concurrent with EGFR-sensitizing mutations (Table 1) [103]. Such mutations are typically found in exons 18–21 of HER2 and alter the tyrosine kinase domain, which activates the receptor in the absence of ligand [103]. The most common HER2 mutation is a 12-bp insertion at exon 20, codon 776, which duplicates the amino acid sequence YVMA (HERYVMA) and thereby alters the HER2 ATP-binding pocket [103]. HERYVMA can activate EGFR in the absence of EGFR ligands to bypass sensitivity to erlotinib and other EGFR-specific TKIs (Table 1) [103]. However, because mutant HER2 compensates for EGFR inhibition in cells that harbor HERYVMA, these cells are instead sensitive to HER2 and dual HER2/EGFR inhibitors, which suggests alternative therapeutic approaches for individuals with NSCLC who harbor mutations in HER2 [103].
Our review focuses on parallel pathways that involve MET, HER2, and HER3; however, other RTKs activate the same downstream signaling cascades and thus could also mediate responses to EGFR inhibition. Namely, the vascular endothelial growth factor receptor (VEGFR), the fibroblast growth factor receptor (FGFR), the platelet-derived growth factor receptor (PDGFR), the AXL/UFO receptor tyrosine kinase, the insulin-like growth factor 1 receptor (IGF1R), and still other RTKs similarly appear to contribute to the observed resistance to EGFR inhibition in NSCLC [105-109]. Nevertheless, our summarized findings highlight the clinical value of the assessment of the genetic variation in RTKs beyond EGFR that have the potential to activate oncogenic pathways despite the presence of EGFR inhibitors. Our findings also emphasize mechanisms of both primary and acquired resistance; specifically, mutations or aberrations in other RTKs can exist concurrently with EGFR mutations and can even be present prior to EGFR-targeted therapy.
Discussion and conclusions
Focused pharmacogenomics research has discovered much about EGFR and how EGFR mutations and expression can guide EGFR-targeted treatment options in patients with NSCLC. The genetic variations in EGFR, however, are unable to accurately predict drug responses to EGFR-targeted agents for all patients with hyperactive EGFR signaling, particularly during treatment with erlotinib and cetuximab (Table 1). In patients with mutant EGFR, for instance, EGFR mutations alone cannot account for all of the observed primary drug resistance in NSCLC patient groups. Therefore, we need to gain a better understanding of why these patients with hyperactive EGFR signaling have such variable responses to EGFR-targeted therapies by designing studies to explore variation in other non-EGFR genes. While we have primarily focused on somatic mutations and the differences in protein expression beyond EGFR that cause primary drug resistance to EGFR-targeted therapies, our findings extend beyond primary drug resistance; namely, the same genetic variation that is detailed in Table 1 might help to explain the rampant acquired resistance to EGFR-targeted agents that quickly follows an initial therapeutic response in a majority of patients with NSCLC who are treated with either erlotinib or cetuximab.
Deciphering the impact of mutations in, and variable expression of, non-EGFR genes on EGFR-targeted NSCLC treatment has significant and immediate clinical applications. At the prescriber level, one potential way to address the variations in other RTKs or in downstream pathway members of EGFR is to implement combination therapy. Given the relatively small number of RTKs in the human genome, more extensive genotyping can be performed to screen for the common activating mutations in non-EGFR RTKs. This would guide EGFR-targeted therapy choices and options for combination therapy. Thorough RTK screening could reveal mutant RTKs with oncogenic phenotypes that are silent or masked by the presence of the more aggressive mutant EGFR. If mutations are found in multiple RTKs, combination therapy approaches that target all identified RTKs could be a powerful way to minimize the risk of acquired resistance to EGFR-targeted therapy.
For instance, EGFR-targeted inhibitors could be coupled with inhibitors of KRAS, MET, PI3K, IKK, HER2, HER3, or even with inhibitors of HER2/HER3 dimerization, to anticipate resistance mediated by genetic abnormalities in members of these downstream or parallel pathways. In fact, several MET inhibitors are in phases II and III clinical trials in the United States, and some are even being examined in combination therapy regimens along with EGFR TKIs such as cetuximab [1,110,111]. The development of additional dual-therapy approaches that investigate the benefits of EGFR-targeted therapy in combination with other RTK inhibitors and downstream pathway inhibitors is in progress, and some combinatorial regimes show clinical promise over EGFR inhibitors alone [112,113]. It is important to note, however, that many of these new drugs are recent discoveries and are still in clinical trials. If and when these drugs are released to the market, the cost, adverse effects, and increased risk of drug-drug interactions versus potential clinical benefit should be taken into account during the process of deciding whether to administer combination therapy in the setting of NSCLC.
Even though the targeting of NSCLC with combination therapy is proving to be a powerful approach in patients whose tumors are resistant to EGFR inhibition, the mixed clinical results indicate that patient responses to combination therapies can be just as varied as the responses to EGFR inhibition. A more comprehensive understanding of the genetic landscape of NSCLC subtypes is thus needed to elucidate modulators of both drug resistance and drug sensitivity to the various combinations of targeted therapies. Therefore, while the pharmacogenomic associations listed in Table 1 provide valuable insight into the treatment of NSCLC, additional genetic factors will inevitably become important in the future as specific, multifaceted therapies are tailored to individual patients.
References
Mayo C, Bertran-Alamillo J, Molina-Vila MA, Gimenez-Capitan A, Costa C, Rosell R. Pharmacogenetics of EGFR in lung cancer: perspectives and clinical applications. Pharmacogenomics. 2012;13:789–802.
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30.
Chi A, Remick S, Tse W. EGFR inhibition in non–small cell lung cancer: current evidence and future directions. Biomark Res. 2013;1:2.
Dziadziuszko R, Jassem J. Epidermal growth factor receptor (EGFR) inhibitors and derived treatments. Ann Oncol. 2012;23:x193–6.
Heuckmann JM, Rauh D, Thomas RK. Epidermal growth factor receptor (EGFR) signaling and covalent EGFR inhibition in lung cancer. J Clin Oncol. 2012;30:3417–20.
Rossi A. Cetuximab and non–small-cell lung cancer: end of the story? Lancet Oncol. 2013;14:1251–3.
Berger C, Madshus IH, Stang E. Cetuximab in combination with anti-human IgG antibodies efficiently down-regulates the EGF receptor by macropinocytosis. Exp Cell Res. 2012;318:2578–91.
Bareschino MA, Schettino C, Troiani T, Martinelli E, Morgillo F, Ciardiello F. Erlotinib in cancer treatment. Ann Oncol. 2007;18:vi35–41.
Passaro A, Alesini D, Pochesci A, Cortesi E. Erlotinib and gefitinib for elderly patients with advanced non–small-cell lung cancer. Anticancer Agents Med Chem. 2014;14:646–50.
Yang X, Yang K, Kuang K. The efficacy and safety of EGFR inhibitor monotherapy in non–small cell lung cancer: a systematic review. Curr Oncol Rep. 2014;16:390.
Douillard JY, Pirker R, O’Byrne KJ, Kerr KM, Storkel S, Von Heydebreck A, et al. Relationship between EGFR expression, EGFR mutation status, and the efficacy of chemotherapy plus cetuximab in FLEX study patients with advanced non–small-cell lung cancer. J Thorac Oncol. 2014;9:717–24.
Shigeta K, Hayashida T, Hoshino Y, Okabayashi K, Endo T, Ishii Y, et al. Expression of epidermal growth factor receptor detected by cetuximab indicates its efficacy to inhibit and proliferation of colorectal cancer cells. PLoS One. 2013;8:e66302.
Pirker R. EGFR-directed monoclonal antibodies in non–small cell lung cancer. Target Oncol. 2013;8:47–53.
Wang Y, Schmid-Bindert G, Zhou C. Erlotinib in the treatment of advanced non–small cell lung cancer: an update for clinicians. Ther Adv Med Oncol. 2012;4:19–29.
Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5.
Califano R, Landi L, Cappuzzo F. Prognostic and predictive value of K-RAS mutations in non–small cell lung cancer. Drugs. 2012;72:28–36.
Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17.
Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21:177–84.
Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34.
Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–7.
Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physio. 2009;218:460–6.
Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;26:641–50.
Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cell Signal. 2005;17:1183–93.
Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, et al. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–69.
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37.
Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–6.
Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203.
Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52.
Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16.
Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006;1:7–9.
Chen Y, Wang BC, Xiao Y. PI3K: a potential therapeutic target for cancer. J Cell Physio. 2012;227:2818–21.
Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71.
Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–96.
Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–9.
Gazdar AF. Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med. 2009;361:1018–20.
Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81.
Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–23.
Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16.
Alarcon T, Page KM. Stochastic models of receptor oligomerization by bivalent ligand. J R Soc Interface. 2006;3:545–59.
Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharm Exp Ther. 2012;343:342–50.
Lim SH, Lee JY, Sun JM, Ahn JS, Park K, Ahn MJ. Comparison of clinical outcomes following gefitinib and erlotinib treatment in non–small-cell lung cancer patients harboring an epidermal growth factor receptor mutation in either exon 19 or 21. J Thorac Oncol. 2014;9:506–11.
Bronte G, Rolfo C, Giovannetti E, Cicero G, Pauwels P, Passiglia F, et al. Are erlotinib and gefitinib interchangeable, opposite or complementary for non–small cell lung cancer treatment? Biological, pharmacological and clinical aspects. Crit Rev Oncol Hemat. 2014;89:300–13.
Cappuzzo F, Ciuleanu T, Stelmakh L, Cicenas S, Szczesna A, Juhasz E, et al. Erlotinib as maintenance treatment in advanced non–small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol. 2010;11:521–9.
Gridelli C, Ciardiello F, Gallo C, Feld R, Butts C, Gebbia V, et al. First-line erlotinib followed by second-line cisplatin-gemcitabine chemotherapy in advanced non–small-cell lung cancer: the TORCH randomized trial. J Clin Oncol. 2012;30:3002–11.
Lee KH, Lee KY, Jeon YJ, Jung MH, Son C, Lee MK, et al. Gefitinib in selected patients with pre-treated non–small-cell lung cancer: results from a phase IV, multicenter, non-randomized study (SELINE). Tuberc Respir Dis. 2012;73:303–11.
Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67.
Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–501.
Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–9.
Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73.
Nebhan C, Pao W. Further advances in genetically informed lung cancer medicine. J Thorac Oncol. 2013;8:521–2.
Pao W, Iafrate AJ, Su Z. Genetically informed lung cancer medicine. J Pathol. 2011;223:230–40.
Keating GM. Afatinib: a review of its use in the treatment of advanced non–small cell lung cancer. Drugs. 2014;74:207–21.
Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–61.
Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–11.
Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–32.
Costa DB, Kobayashi S, Tenen DG, Huberman MS. Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non–small cell lung cancers. Lung Cancer. 2007;58:95–103.
Kobayashi T, Koizumi T, Agatsuma T, Yasuo M, Tsushima K, Kubo K, et al. A phase II trial of erlotinib in patients with EGFR wild-type advanced non–small-cell lung cancer. Cancer Chemother Pharm. 2012;69:1241–6.
Pao W, Girard N. New driver mutations in non–small-cell lung cancer. Lancet Oncol. 2011;12:175–80.
Baty F, Rothschild S, Fruh M, Betticher D, Droge C, Cathomas R, et al. EGFR exon-level biomarkers of the response to bevacizumab/erlotinib in non–small cell lung cancer. PLoS One. 2013;8:e72966.
Pirker R, Pereira JR, Szczesna A, Von Pawel J, Krzakowski M, Ramlau R, et al. Cetuximab plus chemotherapy in patients with advanced non–small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet Oncol. 2009;373:1525–31.
Pirker R, Pereira JR, Von Pawel J, Krzakowski M, Ramlau R, Park K, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non–small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012;13:33–42.
Khambata-Ford S, Harbison C, Woytowitz D, Awad M, Horak C, Xu LA, et al. K-Ras mutation (mut), EGFR-related, and exploratory markers as response predictors of cetuximab in first-line advanced NSCLC: Retrospective analyses of the BMS099 trial. J Clin Oncol. (Meeting Abstracts) 2009;27:no.15S: 8021.
Khambata-Ford S, Harbison CT, Hart LL, Awad M, Xu LA, Horak CE, et al. Analysis of potential predictive markers of cetuximab benefit in BMS099, a phase III study of cetuximab and first-line taxane/carboplatin in advanced non–small-cell lung cancer. J Clin Oncol. 2010;28:918–27.
Kim ES, Neubauer M, Cohn A, Schwartzberg L, Garbo L, Caton J, et al. Docetaxel or pemetrexed with or without cetuximab in recurrent or progressive non–small-cell lung cancer after platinum-based therapy: a phase 3, open-label, randomised trial. Lancet Oncol. 2013;14:1326–36.
Lynch TJ, Patel T, Dreisbach L, McCleod M, Heim WJ, Hermann RC, et al. Cetuximab and first-line taxane/carboplatin chemotherapy in advanced non–small-cell lung cancer: results of the randomized multicenter phase III trial BMS099. J Clin Oncol. 2010;28:911–7.
Zhu CQ, Da Cunha SG, Ding K, Sakurada A, Cutz JC, Liu N, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–75.
Garassino MC, Marabese M, Rusconi P, Rulli E, Martelli O, Farina G, et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non–small-cell lung cancer. Ann Oncol. 2011;22:235–7.
Maus MKH, Grimminger PP, Mack PC, Astrow SH, Stephens C, Zeger G, et al. KRAS mutations in non–small-cell lung cancer and colorectal cancer: implications for EGFR-targeted therapies. Lung Cancer. 2014;83:163–7.
Ferte C, Besse B, Dansin E, Parent F, Buisine MP, Copin MC, et al. Durable responses to Erlotinib despite KRAS mutations in two patients with metastatic lung adenocarcinoma. Ann Oncol. 2010;21:1385–7.
Danenberg KD, Grimminger PP, Mack PC, Danenberg PV, Cooc J, Stephens C, et al. KRAS mutations (MTs) in non–small cell lung cancer (NSCLC) versus colorectal cancer (CRC): implications for cetuximab therapy. J Clin Oncol. 2010;28:10529.
Santini D, Loupakis F, Vincenzi B, Floriani I, Stasi I, Canestrari E, et al. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist. 2008;13:1270–5.
Wang L, Hu H, Pan Y, Wang R, Li Y, Shen L, et al. PIK3CA mutations frequently coexist with EGFR/KRAS mutations in non–small cell lung cancer and suggest poor prognosis in EGFR/KRAS wildtype subgroup. PLoS One. 2014;9:e88291.
Zimmermann S, Peters S. Going beyond EGFR. Ann Oncol. 2012;23:197–203.
Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non–small-cell lung cancer. Nat Rev Cancer. 2010;10:760–74.
Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–9.
Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–706.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62.
D’Amato V, Rosa R, D’Amato C, Formisano L, Marciano R, Nappi L, et al. The dual PI3K/mTOR inhibitor PKI-587 enhances sensitivity to cetuximab in EGFR-resistant human head and neck cancer models. Br J Cancer. 2014;110:2887–95.
Rebucci M, Peixoto P, Dewitte A, Wattez N, De Nuncques MA, Rezvoy N, et al. Mechanisms underlying resistance to cetuximab in the HNSCC cell line: role of AKT inhibition in bypassing this resistance. Int J Oncol. 2011;38:189–200.
Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–61.
Endoh H, Yatabe Y, Kosaka T, Kuwano H, Mitsudomi T. PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol. 2006;1:629–34.
Kim SM, Kim JS, Kim JH, Yun CO, Kim EM, Kim HK, et al. Acquired resistance to cetuximab is mediated by increased PTEN instability and leads cross-resistance to gefitinib in HCC827 NSCLC cells. Cancer Lett. 2010;296:150–9.
Mao C, Liao RY, Chen Q. Loss of PTEN expression predicts resistance to EGFR-targeted monoclonal antibodies in patients with metastatic colorectal cancer. Br J Cancer. 2010;102:940.
Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–30.
Furukawa M, Soh J, Yamamoto H, Ichimura K, Shien K, Maki Y, et al. Silenced expression of NFKBIA in lung adenocarcinoma patients with a never-smoking history. Acta Med Okayama. 2013;67:19–24.
Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–58.
Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–6.
Scartozzi M, Bearzi I, Pierantoni C, Mandolesi A, Loupakis F, Zaniboni A, et al. Nuclear factor-kappaB tumor expression predicts response and survival in irinotecan-refractory metastatic colorectal cancer treated with cetuximab-irinotecan therapy. J Clin Oncol. 2007;25:3930–5.
Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–57.
Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–48.
Sadiq AA, Salgia R. MET as a possible target for non–small-cell lung cancer. J Clin Oncol. 2013;31:1089–96.
Park S, Choi YL, Sung CO, An J, Seo J, Ahn MJ, et al. High MET copy number and MET overexpression: poor outcome in non–small cell lung cancer patients. Histol Histopathol. 2012;27:197–207.
Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–8.
Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–87.
Sgambato A, Casaluce F, Maione P, Rossi A, Rossi E, Napolitano A, et al. The c-Met inhibitors: a new class of drugs in the battle against advanced non–small-cell lung cancer. Curr Pharm Des. 2012;18:6155–68.
Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7.
Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43.
Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658–73.
Song N, Liu S, Zhang J, Liu J, Xu L, Liu Y, et al. Cetuximab-induced MET activation acts as a novel resistance mechanism in colon cancer cells. Int J Mol Sci. 2014;15:5838–51.
Jain A, Penuel E, Mink S, Schmidt J, Hodge A, Favero K, et al. HER kinase axis receptor dimer partner switching occurs in response to EGFR tyrosine kinase inhibition despite failure to block cellular proliferation. Cancer Res. 2010;70:1989–99.
Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–56.
Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–67.
Garrido-Castro AC, Felip E. HER2 driven non–small cell lung cancer (NSCLC): potential therapeutic approaches. Transl Cancer Res. 2013;2:122–7.
Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A, et al. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108:668–75.
Yang F, Tang X, Riquelme E, Behrens C, Nilsson MB, Giri U, et al. Increased VEGFR-2 gene copy is associated with chemoresistance and shorter survival in patients with non–small-cell lung carcinoma who receive adjuvant chemotherapy. Cancer Res. 2011;71:5512–21.
Gadgeel SM, Chen W, Cote ML, Bollig-Fischer A, Land S, Schwartz AG, et al. Fibroblast growth factor receptor 1 amplification in non–small cell lung cancer by quantitative real-time PCR. PLoS One. 2013;8:e79820.
Donnem T, Al-Saad S, Al-Shibli K, Andersen S, Busund LT, Bremnes RM. Prognostic impact of platelet-derived growth factors in non–small cell lung cancer tumor and stromal cells. J Thorac Oncol. 2008;3:963–70.
Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60.
Ludovini V, Flacco A, Bianconi F, Ragusa M, Vannucci J, Bellezza G, et al. Concomitant high gene copy number and protein overexpression of IGF1R and EGFR negatively affect disease-free survival of surgically resected non–small-cell-lung cancer patients. Cancer Chemother Pharm. 2013;71:671–80.
Carter CA, Giaccone G. Treatment of nonsmall cell lung cancer: overcoming the resistance to epidermal growth factor receptor inhibitors. Curr Opin Oncol. 2012;24:123–9.
Scagliotti GV, Novello S, Schiller JH, Hirsh V, Sequist LV, Soria JC, et al. Rationale and design of MARQUEE: a phase III, randomized, double-blind study of tivantinib plus erlotinib versus placebo plus erlotinib in previously treated patients with locally advanced or metastatic, nonsquamous, non–small-cell lung cancer. Clin Lung Cancer. 2012;13:391–5.
Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–400.
Rolfo C, Giovannetti E, Hong DS, Bivona T, Raez LE, Bronte G, et al. Novel therapeutic strategies for patients with NSCLC that do not respond to treatment with EGFR inhibitors. Cancer Treat Rev. 2014;40:990–1004.
Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90.
Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non–small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–93.
Acknowledgements
This study was supported by two NIH/NIGMS grants (No. U01GM61393 and No. K08GM089941), a NIH/NCI grant (No. R21 CA139278), the University of Chicago Cancer Center Support Grant (No. #P30 CA14599), the Breast Cancer SPORE Career Development Award (No. CA125183), and a grant from the National Center for Advancing Translational Sciences of the NIH (No. UL1RR024999). We would also like to thank Mark Applebaum, MD, for his excellent advice on the review topic, especially during the early stages of the project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CD and RSH conceived of the topic and scope of the review. CD and SF conducted the literature review. CD, SF, and RSH wrote the manuscript. RSH supervised the project. All authors read, edited and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Delaney, C., Frank, S. & Huang, R.S. Pharmacogenomics of EGFR-targeted therapies in non–small cell lung cancer: EGFR and beyond. Chin J Cancer 34, 7 (2015). https://doi.org/10.1186/s40880-015-0007-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40880-015-0007-9