Abstract
Actinobacillus equuli subsp. equuli is a member of the family Pasteurellaceae that is a common resident of the oral cavity and alimentary tract of healthy horses. At the same time, it can also cause a fatal septicemia in foals, commonly known as sleepy foal disease or joint ill disease. In addition, A. equuli subsp. equuli has recently been reported to act as a primary pathogen in breeding sows and piglets. To better understand how A. equuli subsp. equuli can cause disease, the genome of the type strain of A. equuli subsp. equuli, ATCC 19392T, was sequenced using the PacBio RSII sequencing system. Its genome is comprised of 2,431,533 bp and is predicted to encode 2,264 proteins and 82 RNAs.
Similar content being viewed by others
Introduction
Actinobacillus equuli subsp. equuli, previously known as ‘Bacillus viscosum-equi’, or ‘Shigella equirulis’, is a common resident of the oral flora of healthy horses, as well as that of the alimentary and genital tracts [1,2]. It has also been reported to be present in other host species such as mice, seemingly without ill effect [3] and on rare occasions, has been transmitted through bite wounds to humans [4]. A. equuli subsp. equuli is the etiological agent of sleepy foal disease, an acute form of fatal septicemia in neonatal foals that may progress to a chronic form, joint ill disease, producing lesions in the kidneys, joints, and lungs [5-8]. Horses with A. equuli infection can present with arthritis, bronchitis, pneumonia, pleuritis, peritonitis, sepsis, endocarditis, pericarditis, nephritis, meningitis, metritis, and abortion [7,9-12]. A. equuli subsp. equuli was previously proposed to act as a secondary pathogen in foals; however, a recent study by Layman and colleagues [13] has revealed that A. equuli subsp. equuli has the potential to act as a primary pathogen given favourable conditions. Recently, it has been reported to also be a primary pathogen in sows and piglets [14,15].
The hemolytic counterpart of this bacterium, A. equuli subsp. haemolyticus , is isolated more frequently from the respiratory tract rather than the oral cavity. It can also cause septicemia and sequelae such as arthritis and meningitis, respiratory tract infections, and mare reproductive loss syndrome [8,10,16].
The similar colonial morphology and biochemical markers and shared 16S rRNA sequences make differentiation of A. equuli from Actinobacillus suis difficult [8]. In addition, little is known about the virulence factors of A. equuli subsp. equuli . To be better able to identify and to improve our understanding of the mechanism of pathogen-host interactions [7], the genome of the type strain A. equuli subsp. equuli strain ATCC 19392 T was sequenced. This strain was isolated from foal blood and deposited in the American Type Culture Collection by the Equine Research Station (New Market, UK) in 1953 [17].
Organism information
Classification and features
As a member of the genus Actinobacillus , A. equuli subsp . equuli belongs to the family Pasteurellaceae, class Gammaproteobacteria [18] (Table 1). Phylogenetic analysis using 16S rRNA sequences suggests that A. equuli subsp . equuli is most closely related to A. suis and A. hominis (Figure 1).
Phylogenetic tree based on 16S rRNA sequences of Actinobacillus sensu stricto species plus A. capsulatus and H. parasuis as outgroups. A. equuli subsp. equuli is indicated in bold. The RDP aligner, which applies the Jukes-Cantor corrected distance model to align sequences, and the RDP Tree Builder, which implements the Weighbor algorithm [36] for tree construction were used. Tree building also involved a bootstrapping process in which the values to the left of the branches illustrate the frequency of occurrence of a branch in 100 replicates [37].
A. equuli subsp. equuli is a small, Gram-negative, nonmotile, pleomorphic bacterium [15,16,19] (Figure 2). It is NAD-independent, nonhemolytic, and CAMP negative [15,20]. A. equuli subsp. equuli produces large amounts of extracellular slime that imparts sticky properties in solid and liquid media cultures [19,31]. On nutrient or blood agar, smooth, grayish-white, circular colonies are produced with an average diameter of 1-2 mm after growth for 24 h [35] (Figure 3). On initial culture from clinical material, colonies are viscous and usually rough but become smooth in successive subcultures [1,19]. Growth using liquid culturing methods has been reported to increase viability in comparison to solid media cultures, and viscosity is retained upon repeated subculturing [1,19]. The usual temperature range for growth of this bacterium is 20-39°C, with an optimum at 37°C, though some A. equuli subsp. equuli strains have been shown to grow at temperatures as high as 44°C [33]. Acid but not gas is produced from sucrose, mannitol, galactose, lactose, maltose, mannose, melibiose, trehalose, raffinose, and glycerol fermentation [19,20,33]. A. equuli subsp. equuli is capable of reducing nitrate and produces α-galactosidase, α-glucosidase, β-xylosidase, urease, and oxidase [19,20,33].
Genome sequencing information
Genome project history
A. equuli subsp. equuli was selected for sequencing because of its importance to the horse industry as the etiologic agent of sleepy foal disease and joint ill disease [7]. Sequencing was done at the McGill University and Génome Québec Innovation Centre (Montréal, QC, Canada) using the PacBio RS II DNA Sequencing System, and assembled using PacBio RS II software and Celera Assembler. A. equuli subsp. equuli was annotated using the NCBI Prokaryotic Genome Annotation Pipeline. A summary of the project information and the Minimum Information about a Genomic Sequence is shown in Table 2 [38].
Growth conditions and genomic DNA preparation
A. equuli subsp. equuli was grown from a frozen (-70°C) seed stock on sheep blood agar plates overnight in an atmosphere of 5% CO2 at 37°C. After subculture, well-isolated colonies were used for genomic DNA isolation. Cells were lysed using modified B1 (150 mM Tris · Cl, 50 mM EDTA, 0.5% Tween®-20, 0.5% Triton X-100, pH 8.0) and B2 (750 mM NaCl, 50 mM MOPS, 15% isopropanol, 0.15% Triton X-100, pH 7.0) buffers. DNA was then column purified using a QIAGEN Plasmid Midi Kit (Qiagen, Germany) following manufacturer's protocol for binding and elution. The resultant DNA preparation was characterized using a NanoDrop model ND1000 Spectrophotometer and was diluted to a concentration of ~0.47 mg/μl.
Genome sequencing and assembly
Single Molecule, Real-Time DNA sequencing (Pacific Biosciences) [39] was done to obtain the genome sequence of the A. equuli subsp. equuli ATCC 19392 T. A total of 133,616 raw subreads were generated with an average length of 4,348 bp using two SMRT Cells in a PacBio RSII sequencer. The resultant subread length cutoff value, 29.42, was used in the Basic Local Alignment with Successive Refinement step [40] where short reads were used to correct for errors on long reads [39]. The corrected reads were assembled into contigs according to the Hierarchical Genome Assembly Process (HGAP) workflow using the Celera Assembler and refined using BLASR to align raw reads on contigs [39]. Final processing was conducted using Quiver, a variant calling algorithm, to generate high quality consensus sequences [39]. There were a total of 4,777 corrected reads with an average length of 7,804 bp and a final product of one contig.
Genome annotation
Genes were identified using the NCBI Prokaryotic Genome Annotation Pipeline. The prediction software, GeneMark, is integrated into the pipeline and performs unsupervised gene finding using heuristic Markov Models [41]. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes (IMG) platform [42] developed by the Joint Genome Institute [43] (Table 3).
Genome properties
The genome of A. equuli subsp. equuli is a single circular chromosome that is 2,431,533 bp in length with a G + C content of approximately 40.3%. It is predicted to contain 2,264 genes, of which 2,182 code for proteins and 82 for RNA; 11 pseudogenes are also present (Table 3 and Figure 3). Approximately 3/4 of the predicted genes can be assigned to one of 25 functional COG categories (Table 4). Of particular note with regard to virulence are several lipopolysaccharide genes predicted to encode biosynthetic enzymes for the O-antigen and lipid A components. Adhesins of different types were observed including several autotransporters; a tight adherence locus; prepilins, and fimbriae; a filamentous hemagglutinin homolog was also detected. In addition, several putative iron acquisition systems are present including those for siderophores, hemoglobin and transferrin. A number of toxin and hemolysin genes were also identified including an aqxCABD operon, although compared to the aqxCABD of A. equuli subsp. haemolyticus there are many point mutations and sizable deletions at both ends of the aqxA gene. Other regions of particular interest include an integron and Mu-like phage, identified using PHAST [44].
Insights from the genome sequence
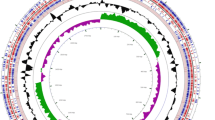
Given the marked similarities of A. equuli and A. suis there has been some debate as to whether these organisms should be a single species. In the current study we determined that the A. equuli subsp. equuli 16S genes are 99% identical to those of both A. suis H91-0380 and the A. suis type strain, ATCC 33415, consistent with membership in the same species. Further, as can be seen in the circular maps below, the genome of A. equuli subsp. equuli is very similar to that of A. suis again suggesting that A. equuli subsp . equuli and A. suis might be the same species (Figure 4). On the other hand, when genomes of A. suis H91-0380 and A. suis ATCC 33415 were compared with that of A. equuli subsp. equuli using the ANI calculator [45], the ANI value of both comparisons was 93.82%, which is lower than 95%, the recommended cutoff value for delineating species [46].
Circular map of the A. equuli subsp. equuli ATCC 19392T genome generated using the CGView Server [49]. From the outside to the center: coding sequences (CDSs) in positive strand, reverse strand CDSs, BLASTN versus A. suis strain H91-0380 (CP003875), BLASTN versus A. suis ATCC 33415 (CP009159), GC content, and GC skew.
In-silico DNA-DNA hybridization, done using a Genome Blast Distance Phylogeny approach to generate genome based distance measures for phylogenetic inferences, also demonstrated differences between A. equuli and A. suis. The Genome-to-Genome Distance Calculator [47] revealed a distance of 0.0685 between A. suis H91-0380 and A. equuli subsp. equuli , with a DDH estimate of 51.40% +/- 2.66. A DDH similarity below 70% is interpreted as two species being distinct; 79% is used to discriminate between subspecies [48]. The DDH estimate exceeding the 70% species threshold was determined from logistic regression to be 23.14%. In terms of subspecies relatedness, the probability of exceeding the 79% threshold was 4.82% between A. equuli subsp. equuli and A. suis H91-0380. The distance calculated between A. suis ATCC 33415 and A. equuli subsp. equuli and their DDH estimate was 0.0681 and 51.60% +/- 2.66, respectively. The probability that DDH exceeded 70% and 79% for A. suis ATCC 33415 and A. equuli subsp. equuli were 23.66% and 4.94%, respectively.
Taken together, these analyses are consistent with the notion that A. suis and A. equuli subsp. equuli are related but distinct species, and care is needed to correctly identify them.
Conclusions
A. equuli subsp. equuli can induce fatal septicemia in foals resulting in significant economic losses in the equine industry; as well, A. equuli subsp. equuli has recently been reported to cause septicemia in swine of all ages. Our analysis of the A. equuli subsp. equuli genome indicates that A. suis and A. equuli subsp. equuli are closely related yet distinct species. At the present time little is known about how A. equuli subsp. equuli causes disease or the factors that control species and tissue tropism. More research including biological experiments is required to better understand the pathogenesis of A. equuli and it is hoped this reported genome sequence of A. equuli subsp. equuli ATCC 19392 T will provide vital information for such studies. In addition, pathway analysis and genome studies may help improve our understanding of host-pathogen interactions of A. equuli subsp. equuli and other Actinobacillus species and aid in the design of diagnostic tools and antimicrobial agents.
References
Edwards PR. Studies on Shigella equirulis. Kentucky Agric Exp Stn Bull. 1931;320:289–330.
Sternberg S, Brändström B. Biochemical fingerprinting and ribotyping of isolates of Actinobacillus equuli from healthy and diseased horses. Vet Microbiol. 1999;66:53–65. PubMed.
Lentsch RH, Wagner JE. Isolation of Actinobacillus lignieresii and Actinobacillus equuli from laboratory rodents. J Clin Microbiol. 1980;12:351–4. PubMed.
Ashhurst-Smith C, Norton R, Thoreau W, Peel MM. Actinobacillus equuli Septicemia: an Unusual Zoonotic Infection. J Clin Microbiol. 1998;36:2789–91. PubMed.
Blackall PJ, Christensen JP, Bisgaard M. Diversity among isolates of Actinobacillus equuli and related organisms as revealed by ribotyping. Aust Vet J. 1998;76:423–5. PubMed.
Aalbaek B, Ostergaard S, Buhl R, Jensen HE, Christensen H, Bisgaard M. Actinobacillus equuli subsp. equuli associated with equine valvular endocarditis. APMIS. 2007;115:1437–42. PubMed.
Berthoud H, Frey J, Kuhnert P. Characterization of Aqx and its operon: the hemolytic RTX determinant of Actinobacillus equuli. Vet Microbiol. 2002;87:159–74. PubMed.
Kuhnert P, Berthoud H, Straub R, Frey J. Host cell specific activity of RTX toxins from haemolytic Actinobacillus equuli and Actinobacillus suis. Vet Microbiol. 2003;92:161–7. PubMed.
Sternberg S. Isolation of Actinobacillus equuli from the oral cavity of healthy horses and comparison of isolates by restriction enzyme digestion and Pulsed-Field Gel Electrophoresis. Vet Microbiol. 1998;59:147–56. PubMed.
Pusterla N, Jones MEB, Mohr RF, Higgins JK, Mapes S, Jang S, et al. Fatal Pulmonary Hemorrhage Associated with RTX Toxin-Producing Actinobacillus equuli subspecies haemolyticus Infection in an Adult Horse. J Vet Diagnostic Investig. 2008;20:118–21. PubMed http://dx.doi.org/10.1177/104063870802000127.
Matthews S, Dart AJ, Dowling BA, Hodgson JL. Peritonitis associated with Actinobacillus equuli in horses : 51 cases. Aust Vet J. 2001;79:536–9. PubMed.
Patterson-Kane JC, Donahue JM, Harrison LR. Septicemia and Peritonitis Due to Actinobacillus equuli Infection in an Adult Horse. Vet Pathol. 2001;38:230–2. PubMed http://dx.doi.org/10.1354/vp.38-2-230.
Layman QD, Rezabek GB, Ramachandran A, Love BC, Confer AW. A retrospective study of equine actinobacillosis cases: 1999-2011. J Vet Diagn Invest. 2014;26:365–75. PubMed http://dx.doi.org/10.1177/1040638714531766.
Thompson AB, Postey RC, Snider T, Pasma T. Actinobacillus equuli as a primary pathogen in breeding sows and piglets. Can Vet J. 2010;51:1223–5. PubMed.
Benavente CE, Fuentealba IC. Actinobacillus suis and Actinobacillus equuli, emergent pathogens of septic embolic nephritis, a new challenge for the swine industry. Arch Med Vet. 2012;44:99–107. http://dx.doi.org/10.4067/S0301-732X2012000200002.
Castagnetti C, Rossi M, Parmeggiani F, Zanoni RG, Pirrone A, Mariella J. Facial cellulitis due to Actinobacillus equuli infection in a neonatal foal. Vet Rec. 2008;162:347–9. PubMed http://dx.doi.org/10.1136/vr.162.11.347.
Public Health England Culture Collections Database. [http://www.phe-culturecollections.org.uk/]
Dewhirst FE, Paster BJ, Olsen I, Fraser GJ. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J Bacteriol. 1992;174:2002–13. PubMed.
Olsen I, Moller K. Genus II. Actinobacillus. In: Garrity G, Brenner D, Krieg N, Staley J, editors. Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume Two, The Proteobacteria, Part B. Second. New York: Springer; 2005. p. 866–83.
Christensen H, Bisgaard M, Olsen JE. Reclassification of equine isolates previously reported as Actinobacillus equuli, variants of A. equuli, Actinobacillus suis or Bisgaard taxon 11 and proposal of A. equuli subsp. equuli subsp. nov. and A. equuli subsp. haemolyticus subsp. nov. Int J Syst Evol Microbiol. 2002;52:1569–76. PubMed http://dx.doi.org/10.1099/ijs.0.01637-0.
Woese CR, Kandlert O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. PubMed.
Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1.
List Editor. Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 2005;55:2235–2238. http://dx.doi.org/10.1099/ijs.0.64108-0
Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1.
Garrity GM, Bell JA, Lilburn T. Pasteurellales ord. nov. In: Garrity GM, Bell JA, Lilburn T. Order XIV. Pasteurellales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 850
List Editor. Validation List no. 7. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1981; 55:382–383. http://dx.doi.org/10.1099/00207713-31-3-382
Pohl SPD. Dissertation, Phillips-Universität Marburg. 1979.
Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol. 1980;30:225–420. http://dx.doi.org/10.1099/00207713-30-1-225.
Brumpt E. Précis de Parasitologie. 1st ed. Paris: Masson et Cie; 1910.
van Straaten H. Bacteriologische bevindingen bij eenigo gevallen van pyo-septicaemie (Lähme) der veuluens. Verslag van den Werksaamheden der Rijksseruminrichting voor 1916-1917, Rotterdam, 1918; 71-76.
Haupt H. Archiv fur wissenschaftliche und praktische Tierheilkunde. 1934; 67:513-524.
Ward CL, Wood JL, Houghton SB, Mumford JA, Chanter N. Actinobacillus and Pasteurella species isolated from horses with lower airway disease. Vet Rec. 1998;143:277–9. PubMed.
MacInnes JI, Lally ET. The Genus Actinobacillus. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes, A Handbook on the Biology of Bacteria: Proteobacteria: Gamma Subclass, vol. 6. 3rd ed. New York: Springer; 2006. p. 1094–118.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology : tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. PubMed http://dx.doi.org/10.1038/75556.
Christensen H, Bisgaard M. Revised definition of Actinobacillus sensu stricto isolated from animals. A review with special emphasis on diagnosis. Vet Microbiol. 2004;99:13–30. PubMed http://dx.doi.org/10.1016/j.vetmic.2003.12.002.
Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol. 2000;17:189–97. PubMed.
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The Ribosomal Database Project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–72. PubMed http://dx.doi.org/10.1093/nar/gkl889.
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. PubMed http://dx.doi.org/10.1038/nbt1360.
Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. PubMed http://dx.doi.org/10.1038/nmeth.2474.
Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. PubMed http://dx.doi.org/10.1186/1471-2105-13-238.
Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–8. PubMed.
Integrated Microbial Genome Database. [http://img.jgi.doe.gov/]
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–7. PubMed http://dx.doi.org/10.1093/nar/gkt963.
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–52. PubMed http://dx.doi.org/10.1093/nar/gkr485.
Average Nucleotide Identity Database. [http://enve-omics.ce.gatech.edu/ani/]
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. PubMed http://dx.doi.org/10.1099/ijs.0.64483-0.
Genome-to-Genome Distance Calculator. [http://ggdc.dsmz.de/distcalc2.php]
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:1–14. PubMed http://dx.doi.org/10.1186/1471-2105-14-60.
Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–4. PubMed http://dx.doi.org/10.1093/nar/gkn179.
Acknowledgements
The authors thank Glenn Soltes for expert technical support. This work was supported by a Natural Sciences and Engineering Research Council to JM; ARB was supported by the Ontario Veterinary College and the Ontario Graduate Scholarship programs.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JIM and AMK contributed to the conception and design of this project. BFH and AMK were involved in the acquisition and initial analysis of the data; BFH, AMK, ARB and JIM were involved in the interpretation of the data. BFH prepared the first draft of the manuscript. All authors were involved in its critical revision and have given final approval of the version to be published and agree to be accountable for all aspects of the work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Huang, B.F., Kropinski, A.M., Bujold, A.R. et al. Complete genome sequence of Actinobacillus equuli subspecies equuli ATCC 19392T . Stand in Genomic Sci 10, 32 (2015). https://doi.org/10.1186/s40793-015-0009-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40793-015-0009-x