Abstract
Background
Multicentre clinical trials evaluating the role of 18F-Fluoroethyl-l-tyrosine (18F-FET) PET as a diagnostic biomarker in glioma management have highlighted a need for standardised methods of data analysis. 18F-FET uptake normalised against background in the contralateral brain is a standard imaging technique to delineate the biological tumour volume (BTV). Quantitative analysis of 18F-FET PET images requires a consistent and robust background activity. Currently, defining background activity involves the manual selection of an arbitrary region of interest, a process that is subject to large variability. This study aims to eliminate methodological errors in background activity definition through the introduction of a semiautomated method for region of interest selection. A new method for background activity definition, involving the semiautomated generation of mirror-image (MI) reference regions, was compared with the current state-of-the-art method, involving manually drawing crescent-shape (gCS) reference regions. The MI and gCS methods were tested by measuring values of background activity and resulting BTV of 18F-FET PET scans of ten patients with recurrent glioblastoma multiforme generated from inputs provided by seven readers. To assess intra-reader variability, each scan was evaluated six times by each reader. Intra- and inter-reader variability in background activity and BTV definition was assessed by means of coefficient of variation.
Results
Compared to the gCS method, the MI method showed significantly lower intra- and inter-reader variability both in background activity and in BTV definition.
Conclusions
The proposed semiautomated MI method minimises intra- and inter-reader variability, providing a valuable approach for standardisation of 18F-FET PET quantitative parameters.
Trial registration ANZCTR, ACTRN12618001346268. Registered 9 August 2018, https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=374253
Similar content being viewed by others
Background
The important role of 18F-Fluoroethyl-l-tyrosine (18F-FET) PET imaging in the management of glioma patients is becoming widely recognised around the world by associations such as the European Association of Neuro Oncology, the European Association of Nuclear Medicine (EANM) and the Response Assessment in Neuro Oncology working group [1,2,3,4]. 18F-FET PET imaging has proven useful in the diagnosis of primary tumour lesions (where biopsy may not be possible), the differentiation between chemoradiation-related changes and tumour recurrence at follow-up, the assessment of response to treatment with certain anticancer drugs and patients’ prognosis [1]. Despite the increasing evidence that 18F-FET PET imaging improves management of glioma patients, there remain critical limitations hampering its inclusion into clinical practice. One barrier to increased use of 18F-FET PET is the prevalence of small single-centre studies or studies based on retrospective PET data collection, which has thus far prevented a robust validation of the clinical value of 18F-FET PET as an imaging biomarker [1]. Consequently, there has been an emerging interest from the neuro-oncology community in combining efforts to validate the utility of 18F-FET PET as an imaging biomarker in prospective multicentre clinical trials, such as the current TROG 18.06 trial [5].
As multicentre trials emerge, there is a need to standardise methods for data acquisition and analysis, to enable correlations and comparisons of results from different sites. Evaluation metrics of such trials are often determined via PET tracer uptake measures, such as standard uptake values (SUV) and tumour-to-brain ratio (TBR), where TBR is defined as the ratio between activity in the tumour lesion and activity in a background reference region in the healthy, contralateral part of the brain. In the current literature on published 18F-FET PET studies, there is large variability in reported values of SUV and TBR taken as threshold for differentiation between tumour and non-tumour tissue [6]. Variability of SUV values between studies arises not only from the acquisition of scans at different timepoints, but also from patients’ physiological factors that can affect 18F-FET SUV in various brain regions, and thereby affect the quantification of 18F-FET uptake in brain tumours [7]. Thus, due to this large intra-patient variability in 18F-FET SUV values, TBR values are a preferred evaluation metric in the intra- and inter-individual comparison of PET results [6]. However, TBR values are also variable in the literature and in clinical practice, and their variability arises from the intra- and inter-reader variability in the selection of the background reference region. Despite the EANM guidelines for brain imaging highlight that the choice of background reference region is a critical step for the reliable quantification of TBR measurements [8], there is still no standard procedural recommendation for this process. Thus, inconsistent approaches are found in the current literature and in clinical practice, mostly involving the arbitrary definition of a 2D circular region of interest or a 3D spherical volume of interest (VOI) in the contralateral hemisphere including grey and white matter [9,10,11,12,13,14,15,16,17]. A recent study by Unterrainer et al. [6] has demonstrated that adding guidelines to the definition of a crescent-shaped VOI in the contralateral hemisphere including grey and white matter significantly reduces intra- and inter-reader variability in the measurement of reference background activity, thus providing a first important step towards the standardisation of background activity assessment for clinical application. However, this method still requires significant manual reader input, which remains time-consuming and a source of methodological error. This variability in reference regions definition is then reflected in the variability in values of background SUV and, consequently, in biological tumour volume (BTV) delineation, which is based on TBR threshold-based segmentation [6]. The resulting uncertainty in BTV definition impacts several therapeutic decision-making processes, ranging from radiotherapy treatment planning to the assessment of treatment response.
In this study, we present a semiautomated method for the generation of background reference regions in the contralateral hemisphere. We show that this semiautomated method improves on gold standard techniques by minimising intra- and inter-reader variability, substantially reducing the time spent by the reader on manual contouring and accounting for tumour size and specific location within the brain. This method encompasses the automated generation of a mirror-image (MI) VOI in the contralateral hemisphere with respect to the anterior–posterior midline reflecting the size, shape and location of the tumour. The strength of this method is that the selection of the size and location of the reference region is not arbitrary, but it reflects the characteristics of the tumour lesion. We aim to assess the efficiency of the MI method by comparing the manual time spent by the reader and the intra- and inter-reader variability in measurements of background SUV and BTV obtained with this method versus that obtained with the guided crescent-shape (gCS) VOI method published by Unterrainer et al. [6]. We selected the gCS as the standard method of comparison because it is the manual method with the lowest intra- and inter-reader variability reported in recent literature [6]. We hypothesize that the MI method will lead to significantly lower values of variability in background mean SUV (SUVmean) and BTV and reduced manual reader input time compared to the gCS method.
Methods
Clinical trial information
This study was a diagnostic, non-randomised, uncontrolled, open-label, single-centre, single-arm, bioavailability, pilot clinical trial, enrolling recurrent glioblastoma multiforme (GBM) patients. Adult patients were eligible for inclusion if they had previously histologically confirmed GBM at resection, progression noted on pre-enrolment MRI scan and an Eastern Cooperative Oncology Group performance status score of ≤ 2. Exclusion criteria were pregnancy, lactation and residence geographically remote from the treating centre. All patients enrolled in the study provided written informed consent in accordance with institutional guidelines. Ethics approval for this study was obtained from the Bellberry Human Research Ethics Committee in August 2018 (Ethics approval number: 2017-11-885). Recruitment for the trial (registration No./date: ACTRN12618001346268/09-08-2018) started in October 2018 and was completed in October 2021, with a total enrolment of ten patients. Details of the data acquisition protocols are available on the trial registration page [18].
Data, readers and manual input objects
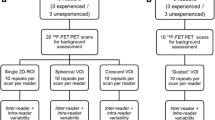
For this study 18F-FET PET data of ten patients with recurrent GBM obtained as part of the Genesis GBM 001 clinical trial were used. Image pre-processing steps included conversion of values of activity into SUV, registration of PET images with CT images, and brain extraction. If severe head rotation was observed in the axial view, a rigid rotation transformation was applied to align the head positions with the image’s orthogonal coordinates before generating brain-extracted image. Complete details of imaging method and image pre-processing steps are included in Additional file 1. Seven readers, including five researchers in medical imaging with at least three years of experience in PET image analysis (C.B., S.P., S.L., P.B., A.G.) and two radiation oncologists (M.F., K.N.), were involved in providing manual inputs for this study. The readers were provided with brain-extracted 18F-FET PET images of the patients in units of SUV and an instruction manual for the generation of the required manual input objects for each of the two methods (MI and gCS). The manual input objects required from each reader on each dataset for the gCS method included the coordinates (x,y,z) of a seed located within the main tumour lesion (region of high 18F-FET PET SUV) and a crescent-shape VOI manually defined according to guidelines as per Unterrainer et al. [6]. Conversely, the manual input objects for the MI method only included the coordinates of a seed located within the main tumour lesion. To enable evaluation of intra-reader variability, each reader provided six repeats of the manual input objects for each patient's dataset.
Segmentation methods
Mirror-image method
The algorithm for the MI method for background contralateral reference (CTRL) VOI definition was developed in Python and is available on GitHub at the following link https://github.com/cbri92/FETsegmenter.git. The 18F-FET PET brain-extracted image and the coordinates of the defined seed within the 18F-FET-enhancing lesion are used to develop an initial segmentation of the 18F-FET-enhancing tumour lesion (BTV0) on the 18F-FET PET image by use of a region-growing algorithm and a FET SUV threshold of 2.2. This value has been previously determined as a cut-off threshold for identification of recurrent glioma [19, 20]. This initial BTV0 is used to automatically generate a MI VOI in the contralateral hemisphere (CTRL0 VOI), excluding potential overlapping areas. The SUVmean calculated in the CTRL0 VOI is then used to normalise the 18F-FET PET image and generate a FET TBR map. The FET TBR map is then used as new input image for growing a new BTV from the input seed coordinates, this time with a FET TBR threshold set at 1.9. This threshold value was chosen based on literature demonstrating the utility of this value in identifying tumour recurrence and progression [21, 22]. The process is repeated in a loop until the convergence condition is reached. The convergence condition set is that the volume of the BTV defined on the FET TBR map equals the volume of the generated CTRL VOI from the previous iteration ± 0.2 cm3. Once convergence is reached, any volume of the CTRL VOI overlapping with the BTV is removed from the CTRL VOI final segmentation, hence excluding infiltrating tumour tissue from the selected background reference region. Then, the SUVmean in the MI CTRLMI VOI and the volume of the BTVMI are extracted for statistical analysis. A schematic representation of the algorithm pipeline is illustrated in Fig. 1. This method accounts for the presence of multiple tumour lesions (defined as multiple seeds by the reader) in the generation of the FET TBR map and, consequently, of the BTVMI.
Guided crescent-shape method
The generation of the CTRL VOI and BTV with the guided crescent-shape method requires as inputs the 18F-FET PET brain-extracted image, the coordinates of the reader-defined seed and the crescent-shape VOI manually drawn by the reader (CTRLCS VOI). First, the SUVmean in the CTRLCS VOI is calculated and used to normalise the 18F-FET PET brain-extracted image to generate a FET TBR map. Then, the segmentation of the BTVCS is developed on the FET TBR map by use of a region-growing algorithm and a FET TBR threshold of 1.9, based on previous studies and consistent with the MI method [21, 22]. For this method no optimisation is performed and the SUVmean in the crescent-shape CTRLCS VOI and the volume of the BTVCS are directly extracted for statistical analysis. A schematic representation of the pipeline used for the crescent-shape method is illustrated in Fig. 1. This method does not account for the presence of multiple tumour lesions in the generation of the FET TBR map and, consequently, of the BTVCS, as FET TBR map generation is solely determined by the manually drawn CTRLCS VOI defined by the reader.
Evaluation metrics
The parameters used to compare the variability between the two studies include the SUVmean in the CTRL VOI and the volume of BTV. For these two parameters variability is assessed by calculating the coefficient of variation (CoV), defined as the ratio between the standard deviation and the mean value of the parameter. Intra-reader variability is defined as the individual CoV of the parameter’s value obtained from the six repeats of a particular scan. Inter-reader variability is defined as the CoV of the mean value of the parameter obtained from the seven readers regarding a particular scan. Intra- and inter-reader reliability are also evaluated via intraclass correlation coefficient (ICC). ICC estimates and their 95% confident intervals are calculated in Python using the pingouin statistical package based on two-way random-effects, absolute-agreement, single rater/measurement model [23]. ICC values < 0.5 indicate poor reliability, values 0.5–0.75 indicate moderate reliability, values 0.75–0.9 indicate good reliability and values > 0.90 indicate excellent reliability [24]. The time taken for the definition of the manual inputs from each reader and the time taken by the algorithm to generate a BTV from the readers’ inputs are also reported for both methods.
Statistical analysis
The statistical analysis performed to assess intra-reader variability between the MI and the gCS methods is a two-tailed, matched-pairs Wilcoxon signed rank test, α = 0.05, between values of CoV for each parameter for each reader. Additionally, overall group comparison on intra-reader variability is assessed by combining the CoV of a parameter from each reader and performing a two-tailed, Mann Whitney unpaired U test, α = 0.05. The statistical analysis performed to assess inter-reader variability between the MI and the gCS methods is a two-tailed, matched-pairs Wilcoxon signed rank test, α = 0.05, between values of CoV for each parameter obtained from all the readers.
Results
Guided crescent-shape VOI
The use of the gCS method for background activity assessment resulted in a median intra-reader CoV of 1.72% (range 0.34–9.99%) for CTRL SUVmean and 6.77% (range 0–65.23%) for BTV, and in a median inter-reader CoV of 2.80% (range 1.00–4.35%) for CTRL SUVmean and 14.37% (range 5.03–36.30%) for BTV (Tables 1 and 2). The average time spent by each reader for the generation of the manual inputs required for this method for a single repeat of one patient was 138 s, and the time taken by the algorithm to generate a BTV from the reader inputs for a single repeat of one patient was 17 s (Table 3).
Mirror-image VOI
The use of the MI method for background activity assessment resulted in a median intra-reader CoV of 0% (range 0–2.15%) for CTRL SUVmean and 0% (range 0–3.88%) for BTV, and in a median inter-reader CoV of 0.005% (range 0–1.05%) for CTRL SUVmean and 0.05% (range 0–36.00%) for BTV (Tables 1 and 2). The time spent by each reader for the generation of the manual inputs required for this method for a single repeat of one patient was 54 s, and the time taken by the algorithm to generate a BTV from the reader inputs for a single repeat of one patient was 54 s (Table 3).
Comparison of background assessment methods
An example of the background reference regions obtained with both methods is shown in Fig. 2. The comparison between MI and gCS methods reveals that all the readers obtained significantly different (p < 0.001) mean values of CTRL SUVmean and BTV for almost all datasets (Additional file 1: Fig. S1). While no clear trend was established, for eight of the ten datasets the gCS method resulted in higher estimates of CTRL SUVmean and, consequently, lower estimates of BTV than the MI method (Additional file 1: Fig. S2). Additionally, for each dataset the mean values of CTRL SUVmean and BTV were more consistent across readers when determined via MI method rather than gCS method (Additional file 1: Fig. S2). When considering variability metrics, all readers obtained significantly lower values of intra- and inter-reader CoV both for CTRL SUVmean (p < 0.0001 and p = 0.002, respectively; Fig. 3 and Additional file 1: Fig. S3) and BTV (p < 0.0001 and p = 0.002, respectively; Fig. 3 and Additional file 1: Fig. S3) by use of MI method. While intra- and inter-reader ICC values revealed excellent reliability in estimates of BTV and CTRL SUVmean with both methods, the MI method resulted in higher ICC values than the gCS (Additional file 1: Fig. S4, Tables S1, S2). The average time spent by each reader in generating inputs for the MI method was ~ 2.5 times faster than for the gCS method (Table 3).
Representative images and segmentations obtained with the two background assessment methods. Top–bottom: 18F-FET PET images of ten GBM patients; 18F-FET TBR images with overlays of BTV (red) and CTRL VOI (green) obtained with the gCS method; 18F-FET TBR images with overlays of BTV (red) and CTRL VOI (green) obtained with the MI method. BTV = biological tumour volume; CTRL = contralateral background reference region; FET = 18F-Fluoroethyl-l-tyrosine; TBR = tumour-to-brain ratio; VOI = volume of interest
Summary plots of intra-reader and inter-reader coefficient of variation (CoV) for the overall group. The plots show the comparison of the intra-reader and inter-reader CoV of the CTRL SUVmean (top row) and BTV (bottom row) between the MI (blue) and the gCS (red) methods for the overall group of readers. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = no significant difference. BTV = biological tumour volume; CoV = coefficient of variation; CTRL = contralateral background reference region; gCS = guided crescent-shape; MI = mirror-image; SUV = standard uptake value
Discussion
The adoption of a consistent method for the standardisation of background activity definition in 18F-FET PET imaging is needed to ensure the reproducible and reliable quantification of 18F-FET uptake parameters, which is necessary for the comparison of multicentre 18F-FET PET clinical trials. Previous studies show that the intra- and inter-reader variability associated with current, most common methods of selecting regions of background activity (2D circular region of interest or 3D spherical VOI) result in background SUV changes of up to ± 8% [6]. This large variability has been mostly attributed to the variable size and insufficient inclusion of different types of tissue within the selected 2D reference region, and to the imprecise and arbitrary selection of the positioning of the 3D reference region, which could result in the inclusion of areas with notably higher 18F-FET uptake, such as venous structures and areas of grey matter [25]. The use of a crescent-shape VOI has been accepted and recommended by the joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards as a solution to these issues, as this method allows for the inclusion in the reference region of a larger volume of tissue from both white and grey matter, and for the morphological adaptation of the reference region such that to exclude ventricles and venous sinuses [26]. Unterrainer et al. [6] were able to demonstrate that when the readers are given a set of rules to draw the crescent-shape VOI, the variability in values of background SUV is significantly reduced compared to the other methods, regardless of the level of experience of the reader performing the task. In our study, we were able to reproduce similar values of intra- and inter-reader CoV for the CTRL SUVmean obtained by use of the gCS method to the values reported by Unterrainer et al. [6], i.e. group intra-reader CoV median 1.10% (range 0.52–2.36%) and group inter-reader CoV median 1.19% (range 0.84–1.89%). However, when we investigated how this variability would propagate in the definition of the BTV, our results demonstrated a substantially higher intra-reader CoV with changes up to 65%. In clinical practice this large dispersion associated with the delineation of BTV can significantly impact several important decision-making processes, such as the ability to assess response to chemotherapy or antiangiogenic therapy, where changes > 20% in BTV are considered treatment response, or in radiotherapy treatment planning, where the BTV is used to delineate areas that should receive a dose-boost [27,28,29,30,31].
In this study, we validated a semiautomated method that could eliminate the variability in background activity assessment, with the aim to also eliminate, or at least minimise, the variability in BTV definition compared to the gCS method. We propose the MI method, which involves the semiautomated generation of a background reference volume as mirror-image of the tumour volume, as a robust method lacking any arbitrary interpretation. Our analysis showed that using the MI method to generate background reference regions not only resulted in a significant reduction in intra- and inter-reader variability in values of CTRL SUVmean compared to the gCS method (with median values of CoV of 0%), but also minimised variability in BTV definition. As mentioned above, this is critically important for a series of clinical decision-making processes that rely on precise BTV estimates, such as assessment of treatment response and radiotherapy treatment planning. The significant reduction in values of CoV, which had median values of 0% for both CTRL SUVmean and BTV, is mainly due to the elimination of biases involved in the individual selection of placement of the background reference region, but also to the consideration of the size and percentage of involvement of different tissue types and brain structures which are reflected in the tumour lesion. Taking into account the size and the relative involvement of different tissue types in the background reference region is particularly important as recent studies have demonstrated that selected normal brain structures have intrinsically high physiological 18F-FET uptake, and that patient-specific factors, such as gender and body mass index, can affect values of 18F-FET uptake in the brain in a patient-specific manner [7, 25]. Consequently, if the tumour is small and located in an area of the brain with intrinsically high 18F-FET uptake, selecting a large background reference VOI from other areas of healthy brain might result in an underestimation of the background reference SUV and, in turn, in an overestimation of TBR values. With the MI method these subjective and local variations in 18F-FET uptake are appropriately normalised. Additionally, the MI method is the first method to include different areas of background activity in the final CTRLMI VOI for cases of patients with multiple tumour lesions located in different areas of the brain. This is not accounted for in current methods that involve the selection of a single contiguous VOI.
A possible concern associated with the use of the MI method is the potential for tumour cells to infiltrate the contralateral brain. This concern has been addressed with the implementation of the algorithm used to generate the CTRLMI VOI, that excludes any area of 18F-FET uptake above background from the final reference region. This solution was developed under the assumption that the potential inclusion of any areas of infiltrating tumour with similar 18F-FET uptake to the surrounding healthy brain tissue in the final CTRLMI VOI would not affect the overall CTRL SUVmean value and BTV definition.
Overall, the MI method represents the first semiautomated method resulting in median 0% variability both on the measurement of values of CTRL SUVmean and BTV. The utility of this method for research applications is clear, as it provides a means not only to evaluate 18F-FET quantitative parameters reliably and reproducibly, thereby facilitating the assessment of multicentre clinical trials, but also to perform objective derivations of imaging features used to build 18F-FET PET-based predictive models. However, this method could also be particularly useful in the clinical setting, where it would be used to semiautomatically generate an initial BTV in a much shorter time and more consistent manner than the gCS method, with the assumption that the resulting BTV would then require minimal final adjustments from the physician. Fully automated approaches for segmentation of solid tumours on PET images have also been shown to reduce inter-reader variability, without, however, being able to provide valid and plausible segmentations of all tumours [32]. This is due to the intrinsic limitation of any fully automated algorithm to account for clinical information absent in the PET image, such as knowledge of patient-specific high uptake regions, which could be incorrectly identified as the tumour location by the algorithm [33]. As such, the clinical consensus recognises that standardised technical approaches for glioma PET imaging procedures need active physician involvement in the segmentation process, particularly for the initial selection of the tumour location and in the review of the final contours [33]. Our MI method satisfies this requirement, with a simple and transparent algorithm that can be integrated into commercial treatment planning systems.
Furthermore, while in this study we validated the use of the MI method for the generation of reproducible 18F-FET quantitative parameters for recurrent GBM patients, the use of this method could be expanded to newly diagnosed GBM cases with the appropriate selection of 18F-FET SUV and TBR threshold values. Finally, the application of this method could be expanded for the standardisation of other types of PET images used in neuro-oncology, such as 18F-FMISO, 18F-FDOPA, 18F-FDG, but also for quantification of PET images used in the diagnosis of other neurodegenerative diseases, such as epilepsy and stroke.
A limitation of this study is that the MI method might not be suitable for patients with tumour lesions heavily involving areas of the brain along the anterior–posterior midline, such as the corpus callosum, patients where tumour growth has severely compromised the anatomical symmetry in the contralateral lobe or patients with multifocal bilateral diseases. While these patients represent only a small percentage (~ 10%) of GBM cases seen in clinical practice, developing an automated method for the generation of representative background reference regions for this subgroup of patients should be the focus of future research work.
Another limitation of this study is the lack of a statistical comparison between expert (nuclear medicine physicists, radiation oncologists) and non-expert readers (medical imaging researchers with experience in PET imaging processing). In this regard, we built on the results published by Unterrainer et al. [6], which demonstrated the lack of significant difference in variability of 18F-FET quantitative parameters derived with the gCS method between expert and unexpert readers.
Finally, a limitation of this study is the inclusion of only ten patients’ datasets, which correspond to the number of datasets available from patients who have enrolled in this trial. While this limitation does not affect measurements of inter-reader variability, it could impact results of intra-reader variability. It is worth mentioning that intra-reader variability does not have as big of an impact on routine clinical decisions as does inter-reader variability, as the segmentation of a single patient’s data is normally done only once by a single physician, but it can be repeated by multiple physicians for credentialing. However, for sake of completeness, we suggest that future studies should validate this method on a larger sample dataset.
Future work should focus on evaluating the use of the MI method for the analysis of prospective multicentre clinical trials foreseeing the recruitment of large sample datasets, such as the current TROG 18.06 trial [5].
Conclusions
In conclusion, this study demonstrated that using the semiautomated method of generating mirror-image VOI in the contralateral hemisphere for the assessment of background activity in 18F-FET PET leads to a fast, reliable and reproducible way of deriving 18F-FET PET quantitative parameters. This method could have critical impact in future 18F-FET PET imaging research studies, as it would provide clinicians and researchers in the field of nuclear medicine with a standardised and robust way of selecting a background reference region, thereby facilitating the comparability of 18F-FET PET studies performed at different centres. Additionally, the MI method could find useful applications in routine clinical practice as a robust tool for the more reproducible definition of BTV segmentations, thus helping clinicians reducing bias in diagnostics evaluations and in making treatment decisions.
Availability of data and materials
The data that support the findings of this study are available from GenesisCare but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of GenesisCare.
Code availability
Code for the generation of the MI and gCS segmentation outputs from reader’s input is publicly available on GitHub at the following link https://github.com/cbri92/FETsegmenter.git.
Abbreviations
- BTV:
-
Biological tumour volume
- CTRL:
-
Background contralateral reference
- CoV:
-
Coefficient of variation
- 18F-FET:
-
18F-Fluoroethyl-l-tyrosine
- GBM:
-
Glioblastoma multiforme
- gCS:
-
Guided crescent-shape
- ICC:
-
Intraclass correlation coefficient
- MI:
-
Mirror-image
- SUV:
-
Standard uptake value
- TBR:
-
Tumour to brain ratio
- VOI:
-
Volume of interest
References
Galldiks N, Lohmann P, Albert NL, Tonn JC, Langen K-J. Current status of PET imaging in neuro-oncology. Neuro Oncol Adv. 2019;1(1):1–11. https://doi.org/10.1093/noajnl/vdz010/5499290.
Albert NL, Weller M, Suchorska B, Galldiks N, Soffietti R, Kim MM, et al. Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016;18(9):1199–208.
Najjar AM, Johnson JM, Schellingerhout D. The emerging role of amino acid PET in neuro-oncology. Bioengineering. 2018;5(4):1–15.
Galldiks N, Niyazi M, Grosu AL, Kocher M, Langen K-J, Law I, et al. Contribution of PET imaging to radiotherapy planning and monitoring in glioma patients—a report of the PET/RANO group. Neuro Oncol. 2021;(February):1–13.
Registry ANZCT. TROG 18.06 Clinical Trial. Prospective, multicentre trial evaluating O-(2-[18F]-fluoroethyl)-l-tyrosine Positron Emission Tomography in Glioblastoma (FET-PET in Glioblastoma) [Internet]. https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=378238.
Unterrainer M, Vettermann F, Brendel M, Holzgreve A, Lifschitz M, Zähringer M, et al. Towards standardisation of 18F-FET PET imaging: do we need a consistent method of background activity assessment? EJNMMI Res. 2017;7(1):48. https://doi.org/10.1186/s13550-017-0295-y.
Verger A, Stegmayr C, Galldiks N, Van Der Gucht A, Lohmann P, Stoffels G, et al. Evaluation of factors influencing 18F-FET uptake in the brain. NeuroImage Clin. 2018;17((June 2017)):491–7.
Vander Borght T, Asenbaum S, Bartenstein P, Halldin C, Kapucu Ö, Van Laere K, et al. EANM procedure guidelines for brain tumour imaging using labelled amino acid analogues. Eur J Nucl Med Mol Imaging. 2006;33(11):1374–80. https://doi.org/10.1007/s00259-006-0206-3.
Koopman T, Verburg N, Schuit RC, Pouwels PJW, Wesseling P, Windhorst AD, et al. Quantification of O-(2-[18F]fluoroethyl)-l-tyrosine kinetics in glioma. EJNMMI Res. 2018;8. https://doi.org/10.1186/s13550-018-0418-0.
Munck Af Rosenschold P, Costa J, Engelholm SA, Lundemann MJ, Law I, Ohlhues L, et al. Impact of [18F]-fluoro-ethyl-tyrosine PET imaging on target definition for radiation therapy of high-grade glioma. Neuro Oncol. 2015;17(5):757–63.
Lohmann P, Herzog H, Rota Kops E, Stoffels G, Judov N, Filss C, et al. Dual-time-point O-(2-[18F]fluoroethyl)-l-tyrosine PET for grading of cerebral gliomas. Eur Radiol. 2015;25(10):3017–24.
Lohmann P, Stoffels G, Ceccon G, Rapp M, Sabel M, Filss CP, et al. Radiation injury vs. recurrent brain metastasis: combining textural feature radiomics analysis and standard parameters may increase 18F-FET PET accuracy without dynamic scans. Eur Radiol. 2017;27(7):2916–27.
Romagna A, Unterrainer M, Schmid-Tannwald C, Brendel M, Tonn JC, Nachbichler SB, et al. Suspected recurrence of brain metastases after focused high dose radiotherapy: Can [18F]FET- PET overcome diagnostic uncertainties? Radiat Oncol. 2016;11(1). https://doi.org/10.1186/s13014-016-0713-8
Lundemann M, Munck af Rosenschöld P, Muhic A, Larsen VA, Poulsen HS, Engelholm SA, et al. Feasibility of multi-parametric PET and MRI for prediction of tumour recurrence in patients with glioblastoma. Eur J Nucl Med Mol Imaging. 2019;46(3):603–13.
Poulsen SH, Urup T, Grunnet K, Christensen IJ, Larsen VA, Jensen ML, et al. The prognostic value of FET PET at radiotherapy planning in newly diagnosed glioblastoma. Eur J Nucl Med Mol Imaging. 2017;44(3):373–81. https://doi.org/10.1007/s00259-016-3494-2.
Moller S, Law I, Munck af Rosenschold P, Costa J, Poulsen HS, Engelholm SA, et al. Prognostic value of 18F-FET PET imaging in re-irradiation of high-grade glioma: results of a phase I clinical trial. Radiother Oncol. 2016;121(1):132–7. https://doi.org/10.1016/j.radonc.2016.08.014.
Pauleit D, Floeth F, Hamacher K, Riemenschneider MJ, Reifenberger G, Müller HW, et al. O-(2-[18F]fluoroethyl)-l-tyrosine PET combined with MRI improves the diagnostic assessment of cerebral gliomas. Brain. 2005;128(3):678–87.
Registry ANZCT. Genesis GBM 001 clinical trial. Genesis GBM 001 Phase I/II study of 68Ga-PSMA as a tumour targeting agent in relapsed glioblastoma. [Internet]. http://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=374253.
Rachinger W, Goetz C, Pöpperl G, Gildehaus FJ, Kreth FW, Holtmannspötter M, et al. Positron emission tomography with O-(2-[18F]fluoroethyl)-l-tyrosine versus magnetic resonance imaging in the diagnosis of recurrent gliomas. Neurosurgery. 2005;57(3):505–11.
Pöpperl G, Götz C, Rachinger W, Gildehaus FJ, Tonn JC, Tatsch K. Value of O-(2-[18F]fluoroethyl)-l-tyrosine PET for the diagnosis of recurrent glioma. Eur J Nucl Med Mol Imaging. 2004;31(11):1464–70.
Galldiks N, Stoffels G, Filss C, Rapp M, Blau T, Tscherpel C, et al. The use of dynamic O-(2–18F-fluoroethyl)-l-tyrosine PET in the diagnosis of patients with progressive and recurrent glioma. Neuro Oncol. 2015;17(9):1293–300.
Galldiks N, Dunkl V, Stoffels G, Hutterer M, Rapp M, Sabel M, et al. Diagnosis of pseudoprogression in patients with glioblastoma using O-(2-[18F]fluoroethyl)-l-tyrosine PET. Eur J Nucl Med Mol Imaging. 2015;42(5):685–95. https://doi.org/10.1007/s00259-014-2959-4.
Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–8. https://doi.org/10.1037/0033-2909.86.2.420.
Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15(2):155–63. https://doi.org/10.1016/j.jcm.2016.02.012.
Fuenfgeld B, Mächler P, Fischer DR, Esposito G, Rushing EJ, Kaufmann PA, et al. Reference values of physiological 18F-FET uptake: Implications for brain tumor discrimination. PLoS ONE. 2020;15(4):1–16.
Law I, Albert NL, Arbizu J, Boellaard R, Drzezga A, Galldiks N, et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18 F] FDG: version 1. 0. 2019;540–57.
Hutterer M, Nowosielski M, Putzer D, Waitz D, Tinkhauser G, Kostron H, et al. O-(2–18F-fluoroethyl)-l-tyrosine PET predicts failure of antiangiogenic treatment in patients with recurrent high-grade glioma. J Nucl Med. 2011;52(6):856–64.
Galldiks N, Rapp M, Stoffels G, Fink GR, Shah NJ, Coenen HH, et al. Response assessment of bevacizumab in patients with recurrent malignant glioma using [18F]Fluoroethyl-l-tyrosine PET in comparison to MRI. Eur J Nucl Med Mol Imaging. 2013;40(1):22–33.
Unterrainer M, Suchorska B, Biczok A, Bartenstein P, Tonn J-C, Kreth F-W, et al. Value of 18F-FET PET for chemotherapy monitoring in non-contrast enhancing gliomas. J Nucl Med [Internet]. 2016;57(supplement 2):14 LP – 14.
Piroth MD, Pinkawa M, Holy R, Klotz J, Schaar S, Stoffels G, et al. Integrated boost IMRT with FET-PET-adapted local dose escalation in glioblastomas: Results of a prospective phase II study. Strahlentherapie und Onkol. 2012;188(4):334–9.
Piroth MD, Galldiks N, Pinkawa M, Holy R, Stoffels G, Ermert J, et al. Relapse patterns after radiochemotherapy of glioblastoma with FET PET-guided boost irradiation and simulation to optimize radiation target volume. Radiat Oncol. 2016;11(1):1–9. https://doi.org/10.1186/s13014-016-0665-z.
Pfaehler E, Burggraaff C, Kramer G, Zijlstra J, Hoekstra OS, Jalving M, et al. PET segmentation of bulky tumors: strategies and workflows to improve inter-observer variability. PLoS ONE. 2020;15(3):1–18.
Hatt M, Lee JA, Schmidtlein CR, Naqa I El, Caldwell C, De Bernardi E, et al. Classification and evaluation strategies of auto-segmentation approaches for PET: report of AAPM task group No. 211. Med Phys. 2017;44(6):e1–42.
Acknowledgements
The authors wish to thank The Rotary Club of Rockhampton North for funding the imaging component of the study and GenesisCare Research (especially Vicki Sproule and Sophie Mepham) for assistance in conducting the trial. The authors thank Eng-Siew Koh, Andrew Scott, Martin Ebert, Roslyn Francis, Michael Back, and Dale Bailey for fruitful discussions.
Funding
This study was funded by The Rotary Club of Rockhampton North and GenesisCare Newcastle. This work was supported by the National Health and Medical Research Council (Grant numbers: APP1132471 and APP1194004), the CSIRO Probing Biosystems Future Science Platform and the Cancer Institute NSW (Grant number: 2019/ECF1015).
Author information
Authors and Affiliations
Contributions
CB conceived the design of the study, contributed reader input data, performed data and statistical analysis, and drafted the manuscript. SP and MF participated in the design of the study, contributed reader input data and commented on draft of manuscript. SL, PB, AG and KN contributed reader input data and commented on draft of manuscript. PK and DW participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. All patients enrolled in the study provided written informed consent in accordance with institutional guidelines. Approval was granted by the Bellberry Human Research Ethics Committee (date: 02/08/2018; No.: 2017-11-885).
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplemental methods of image acquisition and pre-processing, and supplemental figures and tables of results of comparison of background assessment methods.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brighi, C., Puttick, S., Li, S. et al. A novel semiautomated method for background activity and biological tumour volume definition to improve standardisation of 18F-FET PET imaging in glioblastoma. EJNMMI Phys 9, 9 (2022). https://doi.org/10.1186/s40658-022-00438-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40658-022-00438-2