Abstract
The Dimensionality Curse is one of the most critical issues that are hindering faster evolution in several fields broadly, and in bioinformatics distinctively. To counter this curse, a conglomerate solution is needed. Among the renowned techniques that proved efficacy, the scaling-based dimensionality reduction techniques are the most prevalent. To insure improved performance and productivity, horizontal scaling functions are combined with Particle Swarm Optimization (PSO) based computational techniques. Optimization algorithms are an interesting substitute to traditional feature selection methods that are both efficient and relatively easier to scale. Particle Swarm Optimization (PSO) is an iterative search algorithm that has proved to achieve excellent results for feature selection problems. In this paper, a composite Spark Distributed approach to feature selection that combines an integrative feature selection algorithm using Binary Particle Swarm Optimization (BPSO) with Particle Swarm Optimization (PSO) algorithm for cancer prognosis is proposed; hence Spark Distributed Particle Swarm Optimization (SDPSO) approach. The effectiveness of the proposed approach is demonstrated using five benchmark genomic datasets as well as a comparative study with four state of the art methods. Compared with the four methods, the proposed approach yields the best in average of purity ranging from 0.78 to 0.97 and F-measure ranging from 0.75 to 0.96.
Similar content being viewed by others
Introduction
Deep Sequencing is the process of DNA fractioning, which dramatically transformed the genomic research field. The advancement that this process witnessed during the last decade has led to the continuous generation of immense amounts of data, putting the genomic field among the top big data generating fields [1]. Although the captured sequence itself does not express ready to use information, it can be transformed throughout a complex process that deduces the proteins drawn from the sequence. In order to predict the expression of this protein and whether it is cancerous or not, the draft genome is compared to previously known cancer genome sequences [2].
The accumulation of genomic data has raised multiple challenges to produce a logical and coherent picture of the genomic basis of Cancer. Although cancer prognosis is very complicated due to the nature of the genomic datasets that contain thousands of features but relatively fewer samples. Early diagnosis is the key to increase the chances of healing, which makes the process highly crucial. Traditional machine learning techniques fall short in this area since they are used to dealing with datasets that have few features and multiple samples [3] leading to the necessity of novel technologies, hence, big data analytical techniques.
Mining genomic data is a challenging process due to the fact that this type of data meets the criteria, problems and challenges of big data. Big data refers to information with massive volume and high dimensional space [4], it is usually defined through its four characteristics: volume (physical size of data); variety (structure and diversity of data types); velocity (rate at which data is being generated and at which it needs to be processed) and value (the usefulness of analyzing big data). To overcome these challenges, frameworks such as the Hadoop’s implementation of MapReduce [5] were developed, which are designed not only to address these issues but also to work with low commodity hardware [6]. Other prominent big data technologies such as Apache Spark [7] support the same applications and share the same parallelization background as Hadoop, while retaining the scalability and fault tolerance of MapReduce with more flexibility. The different new frameworks offer numerous ecosystems that allow data scientists to conduct several operations among the data analysis process. One of the most important steps of this process, when it comes to genomic big data, is the preprocessing task, more precisely the feature selection step due to the complex nature of these datasets. Feature selection is the process of finding a new subset identifying relevant features in the original dataset and discarding irrelevant and redundant features in order to eventually build models in different analysis tasks [8]. Feature selection algorithms can be categorized into six groups (see Table 1): filters, wrappers, embedded, hybrid, ensemble and integrative, depending on the structuration nature of the selection algorithm and the clustering model building [9].
Successfully dealing with big and complex data preferably requires parallel processing and cluster computing. Among the numerous solutions for parallel processing, lately, Apache Spark has proved to be more potent than other solutions when dealing with massive data [15]. “Apache Spark is an open source tool that can complete jobs considerably faster than previous Big Data tools, namely Apache Hadoop, by virtue of its in-memory caching, and optimized query execution” [16]. Spark’s capacity to efficiently and expeditiously process colossal datasets, led us to choose it as a platform for our approach.
The need for parallelizing the feature selection process is highly desired [17], which raises multiple issues due to the complexity of the dependencies between the different features [18]. The usage of efficient feature selection algorithms that ensure high accuracy with time optimization is the key to a successful analysis. Therefore, in this study, a three layered hybrid distributed approach using Apache Spark is proposed and the following contributions were attained:
-
A parallelized version of the BPSO algorithm for effective feature selection.
-
A parallelized combination of PSO and k-means algorithm in order to present a relevant clustering.
-
The approach was tested on five benchmark datasets, breast cancer tumor, Colon cancer, Leukemia, Lung cancer, Gene expression cancer RNA-Seq datasets to prove its efficiency and speed.
-
The SDPSO approach provides an average purity and F-measure scores that are significantly higher than four state of the art methods, namely, k-means, Genetic Algorithm (GA), the Density-Based Spatial Clustering of Applications with Noise (DBSCAN) and the hybrid PSO-GA [19].
The rest of this paper is organized as follows. In “Related works” section, a review of previous related works is presented. “Methods” section highlights the mathematical background, the methods and the components of the proposed approach. The detailed description of the proposed approach is explained in “Our proposed approach” section. “Results” section discusses the experimental design, evaluation metrics, configurations of experiments and a description of the used datasets for the purpose of experimentation, followed by the experimental results. The findings and results of the approach are discussed in “Discussion” section, followed by the limitations of the study in “Limitations of the study” section. In “Conclusion, perspectives and future work” section, the paper is concluded with a summary of the findings and future work directions.
Related works
This section is divided into two main subsections, the first one deals with previously proposed Binary Particle Swarm Optimization (BPSO) based feature selection methods while the second sub section reviews a number of Particle Swarm Optimization (PSO) based approaches for clustering problems (see Tables 2 and 3).
The main idea behind feature selection is choosing subsets of features from an original set. A subset that should necessarily and reasonably represent the original data along with being beneficial for analysis tasks. The feature selection task is centered on the search for an optimal solution in a usually large search space in order to assuage the clustering task. Although immensely critical, a good feature selection algorithm is not sufficient, the later should be supported by an appropriate clustering algorithm. Several researchers have chosen the PSO algorithm for data clustering.
The solutions described above generally focus on one aspect in the analytical process, the proposed approach is a conglomerate one that targets big and complex data through the parallelization of the computation using Apache Spark, as well as combining an integrative feature selection algorithm BPSO with the PSO for clustering. Along with the contribution in enhancing the computational time due to the parallelized implementation, the proposed approach provides the best in average, of purity and F-measure and the lowest entropy when tested with five complex multidimensional datasets compared to four state of the art algorithms.
Methods
Particle Swarm Optimization (PSO) has attracted significant attention as a technique that enhances the feature selection process due to its efficiency in solving optimization problems [30]. In addition for it being an “anytime algorithm” that produces solutions for any given computational time [31], its simple yet effective principle and solid global search capacity, leading to finding the optimal solution in relatively few iterations [28] were among the main motivations behind the choice of the algorithm as the main constituent of our approach. This section provides a description of the background algorithms behind the approach as well as the statistical preliminaries.
Feature selection algorithm
Carefully understanding the dataset along with dimensionality reduction issues before any data analysis process are crucial to the success of the analysis itself [32]. Data preprocessing involves transforming raw data into a format that is suitable for processing. Real-world data is often incomplete, inconsistent, and is likely to contain noise and errors. Data preprocessing is an endorsed method resolving such issues [33]. Feature selection is a preprocessing technique that automatically selects the features which contribute the most either to the clustering process or to the desired output. Having irrelevant features in the dataset can decrease the accuracy of clustering models and forces the clustering algorithm to process based on irrelevant features. Therefore, it is recommended to conduct a feature selection task before training a model. The feature selection algorithm in this work is a PSO-based algorithm termed the Binary PSO algorithm. The algorithm was originally introduced as an optimization technique for real-number spaces and has since then been successfully applied in many areas: function optimization, artificial neural network training, fuzzy system control, and other application problems [34]. Many optimization problems occur in a space featuring discrete, qualitative distinctions between variables and between levels of variables. Kennedy and Eberhart introduced BPSO [35], which can be applied to discrete binary variables.
In BPSO, a particle i is a pair \((X_i, V_i)\) of two n-dimensional vectors \(X_i\) and \(V_i\), where n is the total number of genes in a dataset. \(X_i\) is a binary vector that represents the position of the particle and \(V_i\) is the velocity of the particle. In the PSO algorithm, every solution of a given problem is considered as a particle which is able to move in a search landscape. In a binary space, a particle may move to near corners of a hypercube by flipping various numbers of bits, if a set of genes is selected, a bit takes the value 1, if not, it takes the value 0. An example of the process’ outcomes using an example section of the gene expression data is featured in Tables 4 and 5.
The movement of the particle is expressed by the two following vectors, the particle vector, Eq. (1), and the velocity vector, Eq. (2):
where \(x_i^d \in {0,1}, i = 1,2,\ldots,m\) (m is the total number of particles), \(d = 1, 2,\ldots, n\) (n is the dimension of data).
To update the position vector of a particle, the movement direction and the speed of that particle are defined as follows:
where w is the inertial weight, \(v_i^d\) is the velocity of particle i at dimension d, \(c_1\) and \(c_2\) are acceleration constants, \(r_1\) and \(r_2\) are random values, \(x_i^d\) is the position of particle i at dimension d, \(pbest_i^d\) is the best previous position of the \(i{\text{th}}\) particle, \(gbest_i^d\) is the global best position of all particles.
The next velocity is defined by three components, the current velocity \(v_i^d\), the distance towards the personal best (pbest) and the distance towards the global best (gbest). For the second and third components, the distance to the personal best and the distance to the global best must be defined and each component is multiplied by an acceleration constant c and a random value r to increase or decrease its impact.
Equation (4) is applied to update the position of each particle. The velocity in BPSO indicates the probability of the corresponding element in the position vector taking value 1. A sigmoid function s(\(v_i^d\)) is introduced to transform \(v_i^d\) to the interval of (0, 1).
where \(r_3\) is a generated random value.
The BPSO fitness function is:
where: \(A(X_i) \in [0,1]\) is the leave one cross validation accuracy on the training set using the only genes in \(X_i\), \(R(X_i)\) is the number of selected genes in \(X_i\), M is the total number of genes for each sample, \(w_1\) and \(w_2\) are two priority weights corresponding to the importance of accuracy and the number of selected genes, respectively, \(w_1 \in [0.1, 0.9]\), \(w_2 = 1 - w_1\).
Clustering algorithm
After the preprocessing step, a final step of analysis is conducted based on the PSO algorithm. PSO algorithm is a population based stochastic optimization technique developed by Kennedy and Eberhart [36]. As the name itself asserts, this method draws inspiration from natural life of swarms of birds. It uses the same principle to find the most optimal solution to a problem in the search space as birds do to find their most suitable place in a swarm [37]. PSO shares many similarities with evolutionary computation techniques such as Genetic Algorithms (GA) [38], but it is proved that the PSO algorithm provides faster convergence and finds better solutions when compared to GA. The implementation of PSO is also simple with a higher computational efficiency [39]. The many advantages within this algorithm have led us to choose it for the clustering model.
The clustering task is the main goal of the study. Therefore, a combination of PSO algorithm along with k-means algorithm is suggested in this work. PSO is a population-based stochastic optimization technique, it simulates the social behavior of organisms. This behavior can be described as an automatically and iteratively updated system. In PSO, each single candidate solution can be considered a particle in the search space. Each particle makes use of its own memory and the knowledge gained from the swarm as a whole to find the best solution. All of the particles have fitness values, which are evaluated using a fitness function in order to be optimized. During movement, each particle adjusts its position by changing its velocity according to its own experience and according to the experience of a neighboring particle, thus making use of the best position encountered by itself and its neighbor. Particles move in the problem space following a current of optimum particles. The PSO algorithm consists of three steps, which are repeated until a predefined stopping condition is met [40]:
-
1.
Evaluate the fitness of each particle,
-
2.
Update personal and global best fitness and positions,
-
3.
Update velocity and position of each particle,
Fitness evaluation is conducted through supplying the candidate solution to the objective function. Personal and global best fitness and positions are updated through comparing the newly evaluated fitness against the previous personal and global best fitness, and replacing the best fitness and positions. The velocity of each particle i in the swarm is updated using Eq. (6):
In Eq. (6), \(v_i^d\) is the velocity of particle i at time d, and \(x_i^d\) is the position of particle i at time d. The parameters w, \(c_1\), and \(c_2\) (0 \(\le\) w \(\le\) 1.2, 0 \(\le\) \(c_1\) \(\le\) 2, and 0 \(\le\) \(c_2\) \(\le\) 2) are user-specified coefficients. The values \(r_1\) and \(r_2\) (0 \(\le\) \(r_1\) \(\le\) 1 and 0 \(\le\) \(r_2\) \(\le\) 1) are random values regenerated for each velocity update. The value \(pbest_i^d\) is the personal best candidate solution for particle i at time d, and \(gbest_i^d\) is the swarm’s global best candidate solution at time d.
Each of the three terms of the velocity update equation have different roles in the PSO algorithm. The first term \(w \times v_i^d\) is the inertia component, responsible for keeping the particle moving in the same direction it was originally heading. The value of the inertial coefficient w is typically between 0.8 and 1.2, which can either weaken the particle’s inertia or accelerate the particle in its original direction [41]. Generally, lower values of the inertial coefficient speed up the convergence of the swarm to optima, and higher values of the inertial coefficient encourage exploration of the entire search space.
The second term \(c_1r_1 \times (pbest_i^d - x_i^d)\), labeled the cognitive component, acts as the particle’s memory, causing it to tend to return to the regions of the search space in which it has experienced high personal fitness. The cognitive coefficient \(c_1\) is usually close to 2, and affects the size of the step the particle takes toward its personal best candidate solution \(pbest_i^d\). The third term \(c_2r_2 \times (gbest - x_i^d)\), labeled the social component, causes the particle to move to the best region the swarm has previously found. The social coefficient \(c_2\) is typically close to 2, and represents the size of the step the particle takes toward the global best candidate solution gbest the swarm has found so far.
The random values \(r_1\) in the cognitive component and \(r_2\) in the social component cause these components to have a stochastic influence on the velocity update. This stochastic influence causes each particle to move in a semi-random manner influenced in the directions of the personal best solution of the particle and the global best solution of the swarm. In order to keep the particles from moving too far beyond the search space, a technique called velocity clamping is used to limit the maximum velocity of each particle [40]. For a search space bounded by the range \([-x_{max}, x_{max}]\), velocity clamping limits the velocity to the range \([-v_{max}, v_{max}]\), where \(v_{max} = k \times x_{max}\).
The value k represents a user-specified velocity clamping factor, \(0.1 \le k \le 1.0\). In many optimization tasks, such as the ones proposed in the paper, the search space is not centered around 0 and thus the range \([-x_{max}, x_{max}]\) is not an adequate definition of the search space. In such a case where the search space is bounded by \([x_{min}, x_{max}]\), \(v_{max} = k \times (x_{max} - x_{min})/2\) are defined. Once the velocity for each particle is defined, each particle’s position is updated by applying the new velocity to the particle’s previous position:
This process is repeated until a predefined stopping condition is met. Common stopping conditions include a predefined number of iterations of the PSO algorithm or a predefined target fitness value.
Our proposed approach
Medical datasets tend to be characterized by missing values and noise, therefore, before engaging in the analysis step, datasets are cleaned as a first step. The list of available preprocessing functions includes instance selection. Approaches for instance selection can be applied for reducing the original dataset to a roughly manageable volume, leading to a reduction of the computational resources that are necessary for performing the analysis process. Algorithms of instance selection can also be applied for removing noisy instances, before applying learning algorithms, which is a step that can improve the accuracy in clustering problems [42].
Another important step within the preprocessing task is the feature selection process that consist mainly of three types of methods filter, wrapper and embedded methods. Filter methods act as preprocessors that rank the features wherein the highly ranked features are selected and applied to a predictor. In wrapper methods, the feature selection criterion is the performance of a predictor, which is wrapped in a search algorithm that finds a subset giving the highest predictor performance. Embedded methods include variable selection as part of the training process. Three other feature selection methods inspired from the previously mentioned ones, are the hybrid, ensemble and integrative methods, they also have shown good performance depending on the type of the problem in hand.
For this work, a BPSO based wrapper approach to feature selection is proposed along with mutual information of each pair of features, which determines the relevance and redundancy of the selected feature subset. Mutual information is a nonparametric, model-free method for scoring a set of features. It can be used to spot all relevant features, and to identify groups of features that allow building a valid clustering [43]. BPSO algorithm is used in binary discrete search spaces. BPSO along with the entropy of each group of features, evaluate the relevance and redundancy of the selected feature subset [44]. The BPSO-based wrapper is followed by a hybrid PSO-k-means algorithm to process the previously cleaned and preprocessed data in order to achieve a desired performance.
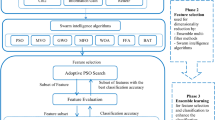
Our SDPSO approach consists of three main phases, (see Fig. 1). The datasets tend to contain null values, extra spaces and insignificant duplications. Therefore, as a first step, datasets are cleaned in order for them to only contain acceptable formats. The cleaned data is then used as an input to construct the BPSO model that provides a preprocessed dataset with a convenient number of features ready for analysis. The approach can be summarized in the following steps:
-
Step 1: Keeping in mind that a particle is considered as the centroid of the cluster, initialize the position and the velocity of the particles.
-
Step 2: Evaluate the fitness for each particle based on Eq. (5).
-
Step 3: If the maximum number of iterations (15 for our approach) is reached go to step 7, if not go to step 4.
-
Step 4: The pbest and the gbest are saved and used to update the particles’ position and velocity according to Eqs. (3) and (4).
-
Step 6: If the gbest is stable when the maximum iterations is reached, move to step 7, if not move back to step 3.
-
Step 7: BPSO results: recommended set of features.
-
Step 8: Reinitialize the swarms from the solution space.
-
Step 9: Evaluate the fitness for each particle.
-
Step 10: The pbest and the gbest are saved and used to update the particles’ position and velocity according to Eqs. (6) and (7). If a particle shows tendency to converge exceeding the boundary, each space is bounded by the range \([-x_{max}, x_{max}]\), velocity clamping limits the velocity to the range \([-v_{max}, v_{max}]\), where \(v_{max} = k \times x_{max}\).
-
Step 11: If the gbest is unvarying when the maximum iterations is reached go to Step 12, if not go back to Step 10.
-
Step 12: Finish the clustering task with the k-means algorithm to find the local optimum using as initial the centroids that resulted from the previous PSO steps.
The BPSO starts by the initialization of the population. First, the position and velocity of particles within the search space are randomly initialized. Second, the fitness values of particles is defined. The first fitness values and positions are the personal best values and the personal best positions (pbest). The global best value and the global best position (gbest) are set to the fitness value and position of the particle with the best fitness value in the entire population. Third, all particles are moved to their new positions using Eq. (3). All fitness values are evaluated again and personal best positions are updated for particles that have a new fitness value (ultimately better than the old personal best value). The global best position is updated if there is any particle with fitness value that is better than the old global best value. All particles are moved to their new positions. The algorithm continues evaluating the fitness values and updating the personal best values, the personal best positions, the global best value, and the global best position. It stops when a limit on the number of iterations is reached.
The clustering algorithm used in the SDPSO is a hybrid one. A combination of PSO with k-means is favored due to the fact that the convergence speed of PSO algorithm near the solution is relatively slow. The k-means algorithm, on the contrary, near the solution, converges fast to a local optimum result, but its ability to find the global solution is weak. Therefore, the combination of the two allows us to find the optimum more quickly. The hybrid algorithm starts with clustering data through PSO, which allows to search all space for a global solution. When the region of global optimum is found by the PSO, the clustering task is resumed using k-means. The hybrid algorithm accelerates the convergence speed as well as the accuracy. Thus, the k-means algorithm finalizes the clustering task. When the value of fitness function for a number of successive iterations changes negligibly the clustering algorithm switches to k-means. All the particles are updated and a new generation of particles is generated. The new particles are used to search the global best position in the solution space. The novelty is that the k-means algorithm is used to search around the global optimum.
Results
Experimental design
In order to test the performance of the SDPSO approach and its capacity to process highly dimensional datasets in low computational runtime, the experimentation is initiated using a non-distributed architecture, followed by a distributed one using PySpark on Spark 2.4. The non-distributed experiments are conducted on a single machine with Ubuntu 16.04 using 4 GB RAM, 4 CPU, and a stable version of Python 2.7 is used to implement the approach. The parallelized experiments are conducted using Apache Spark.
Our approach is implemented through the use of the many advantages presented by Apache Spark, namely, the parallel processing notion using Spark Resilient Distributed Datasets (RDD). In the master node, each particle is modelled through the use of a Python class. A swarm is then created using instances of the class and processed into the RDD. The updating of the particles’ position and velocity are conducted in the slave nodes. The updated best values are sent back to the master node to determine and update the new global best, which is again forwarded to the slave nodes for update. This process continues until the limited number of iterations is met.
Dataset description
Five benchmark datasets are chosen to test the performance of this approach, each dataset has two labels M = Malignant, B = Benign.
-
Breast cancer tumors: A dataset with examples labeled as either malignant or benign with 30 features [45].
-
Colon cancer: A colon cancer dataset which contains information on 62 samples for 2000 genes. The samples belong to tumor and normal colon tissues [46].
-
Leukemia: The total number of genes to be tested is 7129, and the number of samples to be tested is 72 [47].
-
Lung cancer: The lung dataset contains 181 tissue samples. Each sample is described by 12533 genes [48].
-
Gene expression cancer RNA-Seq dataSet: composed of 20531 features, this dataset is used in the experiments due to its complex nature and the diversification of its features [49].
The datasets were selected to have various numbers of features, classes and instances as representative samples of the problems that the proposed approaches can address (see Table 6).
Evaluation metrics
As evaluation metrics to test the performance of our approach, entropy, purity and F-Measure are used. Entropy and purity are widely used measures to determine the clustering efficiency. For each cluster, entropy uses external information class labels to test the performance. Lower entropy means better clustering. The Entropy is magnified when the members of the cluster are more diversified. So we aspire low entropy for every cluster in order to maintain the efficacy of the clustering task. For each cluster, purity determines the largest class and it attempts to capture how well the groups match with the reference on average [50]. To define the entropy E, we start by defining the probability Prob that a member of a cluster j belongs to class i.
where \(N_{ij}\) is the number of members of a class i in a cluster j and \(N_j\) is the number of members in a cluster j. The entropy of a cluster is defined as follows:
where k is the number of classes. The total entropy is defined as follows:
where N is the total of members and n is the number of clusters.
The purity Pu of a cluster is defined as follows:
The overall purity is defined as follows:
where N is the total of members and n is the number of clusters.
F-measure is a technique that combines the precision and the recall measurements from information retrieval literature [51]. The precision P and recall R of a cluster j (generated by the clustering algorithm) with respect to a class i (prior knowledge of the datasets) is defined as [52]:
where \(N_{ij}\) is the number of examples of class i within cluster j, \(N_j\) is the number of items of cluster j and \(N_{i}\) is the number of members of class i. The corresponding value of the F-measure is:
With respect to class i, members of i may be organized into different clusters. That will generate multiple F-measure value for class i. The cluster with the highest F-measure score is considered as the cluster for class i. The overall F-measure for the clustering result of one algorithm is computed as:
where n is the number of the clusters in the dataset and |i| is the number of data objects in class i. The F value is limited within the interval [0, 1]. The higher, the F-measure is, the better the clustering result are.
Experimental results
The experimentations are divided into two main parts. The first experiment tests the BPSO feature selection method against two prominent feature selection methods, which are implemented on the same datasets of interest. The second part of the experimentation is testing the runtime as well as the accuracy of the SDPSO approach. Both experiments are conducted on the five benchmark datasets previously described in “Dataset description” section.
Feature selection results
The preprocessing step using the BPSO-based feature selection algorithm is tested against two prominent feature selection algorithms in order to justify our choice:
-
CFS: Correlation-based feature selection is a filter method that ranks feature subsets according to an appropriate correlation measure and a heuristic search strategy [53].
-
ReliefF: An extension of the binary-classification Relief algorithm [54] which was limited to binary classification problems. ReliefF [55] can deal with multiclass problems, which makes of it an improved algorithm that is more likely to handle incomplete and noisy data [56].
Table 7 displays the number of features selected by each of the three feature selection methods, CFS, ReliefF and BPSO. It is noted that the number of features selected by BPSO is noticeably higher than the number of features selected by CFS and reliefF respectively.
SDPSO results
The accuracy of the SDPSO results, after it was tested on five benchmark datasets with different characteristics are expressed in Table 8 and Fig. 2. Five experimental trials are conducted with each dataset and the means of the five values was taken into consideration as the final result. For Breast cancer and Lung cancer the approach achieved considerably high accuracy. For the rest of the datasets, despite the lower purity and F-Measure scores, it is still in general relatively credited compared to results provided by the four state of the art methods, namely, k-means, GA, DBSCAN and hybrid PSO-GA algorithms, applied on the same datasets of interest. As can be observed, the effectiveness of the SDPSO approach is demonstrated as it yields the lowest in average of entropy 0.16 compared to the average entropy of 0.23, 0.22, 0.19 and 0.18 for the k-means, GA, DBSCAN and hybrid PSO-GA algorithms respectively. Our approach also shows a better purity average of 0.88 and F-measure average of 0.87 compared to a purity average of 0.79, 0.77, 0.82 and 0.85 and an F-score average of 0.82, 0.79, 0.84 and 0.85 for the k-means, GA, DBSCAN and hybrid PSO-GA algorithms respectively.
Another test was conducted to test computational cost as well as to emphasize on the importance of the parallelized approach. The parallelized interpretation of the approach is compared with the non-parallelized one. The average total runtime of the SDPSO approach is indicated in Table 9 for both the non-parallelized and the parallelized implementations. Similarly to the previous experimentation, all the experiments are run five times and the parallelized implementation shows faster runtime compared to the non-parallelized one along with a good accuracy average (see Figs. 3 and 4). Evidently, the number of the features influences the total runtime since it adds to the complexity of the computation. For the Gene Expression Cancer, the total runtime exceeded 8 days without showing any iteration result, which prompts us to consider it as an infinite runtime.
Discussion
Nowadays, it is pivotal in many fields to develop scalable approaches that can efficiently optimize processes and solve scalability issues. In the medical field, due to the critical nature of the data, it is crucial to obtain satisfying efficiency with lower time response. The use of horizontal scaling through Apache Spark combined with conglomerate and encompassing computational techniques allow organizations to access strong computational strength in approachable prices compared to the vertical scaling which demands high commodity hardware. The aim of this work is to provide an encompassing fast and accurate approach that can be afforded by medical organization through the implementation of a PSO-based approach in Apache Spark that is famously known for its capacity to optimize the computation cost in comparison with the vertically scalable solution that require the purchase of high commodity hardware.
Performance and accuracy
In the previous section, the experimental results demonstrate that SDPSO provides promising results both in terms of runtime and accuracy. High accuracy is pivotal, especially with medical data where very little room of error is allowed. SDPSO performance was assessed using five genomic datasets, the breast cancer, the colon cancer, the leukemia, the lung cancer and the genetic expression cancer datasets; it noticeably outperforms four state of the art algorithms, namely, k-means, GA, DBSCAN and hybrid PSO-GA yielding a purity that varies from 0.78 to 0.97, an F-measure that varies from 0.75 to 0.96 and an entropy that varies from 0.06 to 0.30 depending on the complexity of the dataset; taking into consideration that the higher the purity and F-measures are and the lower the entropy is, the higher the performance of the clustering task is.
Parallelization, cost and runtime
Moreover, the Apache Spark implementation of the SDPSO approach shows to be much faster than a single node non-parallelized implementation of the same approach. The BPSO selects more features compared to other feature selection methods, which allows it to be designated as a more inclusive method, eventually allowing the accuracy of the approach as a whole to increase. Owing to Apache Spark’s efficient data processing, the SDPSO approach gains computational power. SDPSO manages to provide a satisfying accuracy in a fairly lower computational time compared to non-parallelized approaches. The use of Apache Spark is remarkably cost friendly, it allows the analysis to run smoothly even with low commodity hardware, which is highly required in the medical field in order to satisfy the needs of the unfortunate medical organizations.
Limitations of the study
Our approach is mainly based on PSO due to the adaptability of the algorithm towards different problems simply by modifying or adding fitness functions. This flexibility, along with its efficiency, allow it to be used to operate a wide range of clustering processes. As highlighted through the article, PSO merits to be widely used due to its capability to adapt to large amounts of data, using fitness functions distinctively meeting the needs of the study in hand, and the fact that it preforms an effective global search of the solution space. Despite its numerous advantages, the PSO-based approaches fall short in many aspects. Due to its nature, PSO algorithm relies on a number of hyper parameters that are user-supplied and critical to determine in practice. Methods to test the cohesion and separation measures of the combination of k-means with PSO are important and ought to be addressed in future works, however, they are beyond the scope of this paper as we rely on the results of the entropy, the purity and the F-measure to validate our approach as a whole.
Conclusion, perspectives and future work
In this work a large scale study of the PSO algorithm is presented both as a feature selection and as a clustering method applied to cancer datasets. The performance of the SDPSO approach is systematically evaluated using five datasets, in order to analyze the influence of the multidimensional datasets on the outcomes of the analysis process. The proposed approach yields the best in average of purity ranging from 0.78 to 0.97 and in average of F-measure ranging from 0.75 to 0.96 compared to four state of the art methods, namely, k-means, GA, DBSCAN and hybrid PSO-GA. Although the results vary within the datasets, the general picture provided here helps emphasize on the importance of the combination of the PSO-based computational techniques with the parallelization’s touch that is added through Apache Spark usage for the prognosis tasks on cancer datasets.
Despite the multiple benefits of parallelism, it can be risky if not used properly. Over-parallelism can be malicious to the accuracy of the results; these issues grow in scale, when large and complex datasets are used, the distribution can result in disregarding certain meaningful relationships between features. In the light of the above, the proposed approach successfully contributes to the reduction of runtime while maintaining reasonable accuracy. In a near future, the hope is to contribute in making the literature on large scale data analysis be as mature as the one small scale data. Our main objective is to help ensure accessibility to strong computational creativity, with the minimum of expenses. Given the large dimensionality of the used datasets, further investigation is required regarding clustering in these spaces, which will be the leading perspective of our future work. The efforts towards elucidating this question will most probably involve the use and evaluation of even more elaborated feature selection and clustering algorithms.
Availability of data and materials
Not applicable.
Abbreviations
- PSO:
-
Particle Swarm Optimization
- BPSO:
-
Binary Particle Swarm Optimization
- SDPSO:
-
Spark Distributed Particle Swarm Optimization
- DPC:
-
Density peaks clustering
- DNA:
-
Deoxyribonucleic acid
- kNN:
-
k-nearest neighbors
- DBSCAN:
-
Density-Based Spatial Clustering of Applications with Noise
- SVM:
-
Support vector machine
- LGO:
-
Local and global optimum
- HBPSOSCA:
-
Hybrid Binary Particle Swarm Optimization and Sine Cosine Algorithm
- NBPSO:
-
New Binary Particle Swarm Optimization
- SC-BR-APSO:
-
Subtractive Clustering based Boundary Restricted Adaptive Particle Swarm Optimization
- FCM:
-
Fuzzy C-means
- SA:
-
Simulated annealing
- MPSO:
-
Modified Particle Swarm Optimization
- DPC:
-
Density peaks clustering
- PSO-SA:
-
Particle Swarm Optimization Simulated Annealing
- GA:
-
Genetic Algorithm
- CFS:
-
Correlation based feature selection
- RDD:
-
Resilient distributed datasets
References
Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Childhood Educ Pract Ed 2013;98(6):236-238.
Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet. 2010;19(R2):R188.
Wong TT, Hsu CH. Two-stage classification methods for microarray data. Expert Syst Appl. 2008;34(1):375.
Safhi HM, Frikh B, Hirchoua B, Ouhbi B, Khalil I. Data intelligence in the context of big data: a survey. J Mob Multimedia. 2017;13(1&2):1.
Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. Commun ACM. 2008;51(1):107.
Khawla T, Fatiha M, Azeddine Z, Said N. A blast implementation in Hadoop MapReduce using low cost commodity hardware. Procedia Comput Sci. 2018;127:69.
Zaharia M, Chowdhury M, Franklin MJ, Shenker S, Stoica I. Spark: cluster computing with working sets. HotCloud. 2010;10(10–10):95.
Solorio-Fernández S, Carrasco-Ochoa JA, Martínez-Trinidad JF. A review of unsupervised feature selection methods. Artif Intell Rev. 2020;53(2):907.
Tadist K, Najah S, Nikolov NS, Mrabti F, Zahi A. Feature selection methods and genomic big data: a systematic review. J Big Data. 2019;6(1):79.
Landset S, Khoshgoftaar TM, Richter AN, Hasanin T. A survey of open source tools for machine learning with big data in the Hadoop ecosystem. J Big Data. 2015;2(1):24.
Kushmerick N, Weld DS, Doorenbos R. Wrapper induction for information extraction. Washington: University of Washington; 1997. p. 729–737.
Naseriparsa M, Bidgoli AM, Varaee T. A hybrid feature selection method to improve performance of a group of classification algorithms; 2014. arXiv:1403.2372.
Tsymbal A, Pechenizkiy M, Cunningham P. Diversity in search strategies for ensemble feature selection. Inf Fus. 2005;6(1):83.
Perscheid C, Grasnick B, Uflacker M. Integrative gene selection on gene expression data: providing biological context to traditional approaches. J Integr Bioinform. 2018;16(1):20180064. https://doi.org/10.1515/jib-2018-0064.
Samadi Y, Zbakh M, Tadonki C. Comparative study between Hadoop and Spark based on Hibench benchmarks. In: 2016 2nd International conference on cloud computing technologies and applications (CloudTech). Marrakech, Morocco: IEEE;2016. p. 267–75.
Siddiqa A, Karim A, Gani A. Big data storage technologies: a survey. Frontiers Inf Technol Electronic Eng. 2017;18(8):1040–70.
Eiras-Franco C, Bolón-Canedo V, Ramos S, González-Domínguez J, Alonso-Betanzos A, Tourino J. Multithreaded and Spark parallelization of feature selection filters. J Comput Sci. 2016;17:609.
Last M, Szczepaniak PS, Volkovich Z, Kandel A, editors. Advances in web intelligence and data mining, vol. 23. Berlin: Springer; 2006. p. 295–304.
Patibandla RL, Rao BT, Krishna PS, Maddumala VR. Medical data clustering using particle swarm optimization method. J Crit Rev. 2020;7(6):363.
Chuang LY, Chang HW, Tu CJ, Yang CH. Improved binary PSO for feature selection using gene expression data. Comput Biol Chem. 2008;32(1):29.
Yang CS, Chuang LY, Ke CH, Yang CH. A hybrid feature selection method for microarray classification. In: IAENG International journal of computer science. New York: IEEE; 2008. p. 2093–8.
Ibrahim TNT, Marapan T, Hasim SH, Zainal AF, Abidin NO, Nordin NA. Jaafar HI, Osman K, Ghani ZA, Hussein SFM. A brief analysis of Gravitational Search Algorithm (GSA) publication from 2009 to May 2013. In: International conference recent treads in engineering & technology (ICRET’2014). Romania; 2014. p. 47–57.
Wei J, Zhang R, Yu Z, Hu R, Tang J, Gui C, Yuan Y. A BPSO-SVM algorithm based on memory renewal and enhanced mutation mechanisms for feature selection. Appl Soft Comput. 2017;58:176.
Kumar L, Bharti KK. An improved BPSO algorithm for feature selection. Recent trends in communication, computing, and electronics. Singapore: Springer; 2019. p. 505–13.
Ghorpade-Aher J, Metre VA. PSO based multidimensional data clustering: a survey. Int J Comput Appl. 2014;87(16):41–48.
Niknam T, Amiri B, Olamaei J, Arefi A. An efficient hybrid evolutionary optimization algorithm based on PSO and SA for clustering. J Zhejiang Univ Sci A. 2009;10(4):512.
Dudeja C. Fuzzy-based modified particle swarm optimization algorithm for shortest path problems. Soft Comput. 2019;23(17):8321.
Cai J, Wei H, Yang H, Zhao X. A novel clustering algorithm based on DPC and PSO. IEEE Access. 2020;8:88200.
Mahesa R, Wibowo EP. Optimization of fuzzy c-means clustering using particle swarm optimization in brain tumor image segmentation. J Theor Appl Inf Technol. 2020;98:19.
Koumi F, Aldasht M, Tamimi H. Efficient feature selection using particle swarm optimization: a hybrid filters-wrapper approach. In: 10th International conference on information and communication systems (ICICS). Irbid: IEEE; 11–13 June 2019. p. 122–7.
Sujit PB, Beard R. Multiple UAV path planning using anytime algorithms. In: American control conference. St. Louis: IEEE; 10–12 June 2009. p. 2978–83.
Al-Tashi Q, Abdulkadir SJ, Rais HM, Mirjalili S, Alhussian H. Approaches to multi-objective feature selection: a systematic literature review. IEEE Access. 2020;8:125076.
Famili A, Shen WM, Weber R, Simoudis E. Data preprocessing and intelligent data analysis. Intell Data Anal. 1997;1(1):3.
Dorrah HT, El-Garhy AM, El-Shimy ME. PSO-BELBIC scheme for two-coupled distillation column process. J Adv Res. 2011;2(1):73.
Kennedy J, Eberhart RC. A discrete binary version of the particle swarm algorithm. In: 1997 IEEE International conference on systems, man, and cybernetics. Computational cybernetics and simulation. New York: IEEE; 1997. p. 4104–8.
Marini F, Walczak B. Particle swarm optimization (PSO). A tutorial. Chemom Intell Lab Syst. 1995;149:153–65.
Juneja M, Nagar SK. Particle swarm optimization algorithm and its parameters: a review. In: 2016 International conference on control, computing, communication and materials (ICCCCM). New York: IEEE; 2016. p. 1–5.
Panda S, Padhy NP. Comparison of particle swarm optimization and genetic algorithm for FACTS-based controller design. Appl Soft Comput. 2008;8(4):1418.
Hassan R, Cohanim B, De Weck O, Venter G. A comparison of particle swarm optimization and the genetic algorithm. In: 46th AIAA/ASME/ASCE/AHS/ASC structures, structural dynamics and materials conference; 2005. p. 1897.
Van den Bergh F. An analysis of particle swarm optimizers [Ph. D. thesis]. Pretoria: Natural and Agricultural Science Department, University of Pretoria; 2001.
Shi Y, Eberhart R. A modified particle swarm optimizer. In: 1998 IEEE international conference on evolutionary computation proceedings. IEEE world congress on computational intelligence. New York: IEEE; 1998. p. 69–73.
Liu H, Motoda H, editors. Instance selection and construction for data mining, vol. 608. Berlin: Springer; 2013.
Krier C, François D, Wertz V, Verleysen M. Feature scoring by mutual information for classification of mass spectra. In: Applied artificial intelligence; 2006. p. 557–564.
Kushwaha N, Pant M. Link based BPSO for feature selection in big data text clustering. Future Gen Comput Syst. 2018;82:190.
Street WN, Wolberg WH, Mangasarian OL. Nuclear feature extraction for breast tumor diagnosis. In: Biomedical image processing and biomedical visualization, vol. 1905. International Society for Optics and Photonics; 1993. p. 861–70.
Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci. 1999;96(12):6745.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7. https://doi.org/10.1126/science.286.5439.531.
Gordon GJ, Jensen RV, Hsiao LL, Gullans SR, Blumenstock JE, Ramaswamy S, Richards WG, Sugarbaker DJ, Bueno R. Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 2002;62(17):4963–7.
Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. Cancer Genome Atlas Research Network. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–20. https://doi.org/10.1038/ng.2764.
Sripada SC, Rao MS. Comparison of purity and entropy of k-means clustering and fuzzy c means clustering. Indian J Comput Sci Eng. 2011;2(3):343.
Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061; 2020.
Cui X, Beaver JM, Charles JS, Potok TE. Dimensionality reduction particle swarm algorithm for high dimensional clustering. In: 2008 IEEE swarm intelligence symposium. St. Louis: IEEE; 21-23 Sept 2008. p. 1–6.
Hall MA. Correlation-based feature selection for machine learning. Hamilton: The University of Waikato; 1999.
Kira K, Rendell LA. The feature selection problem: Traditional methods and a new algorithm. In: AAAI'92: Proceedings of the tenth national conference on Artificial intelligence, vol. 2; 1992. p. 129–134.
Kononenko I. Estimating attributes: analysis and extensions of RELIEF. European conference on machine learning. Berlin, Heidelberg: Springer; 1994. p. 171–82.
Robnik-Šikonja M, Kononenko I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach Learn. 2003;53(1–2):23.
Acknowledgements
The authors thank the anonymous reviewers for their helpful suggestions and comments.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All mentioned authors contribute in the elaboration of the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tadist, K., Mrabti, F., Nikolov, N.S. et al. SDPSO: Spark Distributed PSO-based approach for feature selection and cancer disease prognosis. J Big Data 8, 19 (2021). https://doi.org/10.1186/s40537-021-00409-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40537-021-00409-x