Abstract
Background
Rab protein family is the largest subfamily of small G protein family. As one of the most important families in plant, Rab family plays an important role in the process of plant growth and development. So far, the identification of 57 members of the Rab family in Arabidopsis has been completed. In cotton, the relevant family has not been reported.
Results
Here, we identified 87, 169, 136, 80 Rabs in the four sequenced cotton species, G. raimondii (D5), G. hirsutum acc. TM-1 (AD1), G. barbadense acc. 3-79 (AD2) and G. arboreum (A2), respectively. Biological information analysis showed that the number of amino acid is 200–300 aa among Rab family members in G. raimondii and the protein molecular weight is between 20 and 30 kDa, which is consistent with the characterization of the Rab protein itself. 87 GrRabs in G. raimondii are divided into eight groups. In each group, intron numbers and subcellular localization of Rab protein are basically the same. We mapped the distribution of GrRab genes on 13 chromosomes of G. raimondii except two genes. Among the 87 GrRabs in G. raimondii, we identified 60 pairs of GrRabs formed in whole genome duplication. Among all the gene pairs, the Ka/Ks values were less than 1. This indicates that it is the results of the purification selection and will help maintain the conservation of gene in structure and function. Further, 4 of the 87 GrRabs showed tandem duplication. They were GrRabA2a vs GrRabD1a and GrRabA2h vs GrRabD1b respectively. Expression patterns analysis of 169 GhRabs in G. hirsutum acc. TM-1 indicates that most Rab family members play a certain role in different tissues/organs and different growth stages of cotton, implying their potential function in the polar growth of pollen tube, root hair and fiber cell, as well as improving stress and disease tolerance.
Conclusion
The systematic investigation of Rab genes in cotton will lay a foundation for understanding the functional roles of different Rab members in the polar growth and stress tolerance.
Similar content being viewed by others
Background
As the most important cash crop in the world, cotton provides a very important natural fiber for human beings. Therefore, we tried to explore the important genes related to abiotic stress, biotic stress and fiber development from the whole genome of cotton, hoping to improve cotton quality and yield. Rab protein family in cotton is a very large family. According to previous reports, Rab family is not only closely related to plant growth and development, but also plays an important role in stress and disease resistance.
The world’s first Rab gene (YPT1) was discovered in yeast in 1983 (Gallwitz et al. 1983). Four years later, Salminen and Novick demonstrated that Ras (Sec4) is involved in vesicle trafficking (Salminen and Novick 1987). In the same year, Tavitian and his colleagues cloned the homologous gene similar to the SEC4/YPT gene for the first time through cDNA library in rat brain and named ras-like in rat brain (Rab) (Martinez and Goud 1998). In Arabidopsis thaliana, 57 Rab proteins were found and divided into RabA to RabH (Hill and Sylvester 2007). RabA to RabH in A. thaliana correspond to Rab11, Rab2, Rab1, Rab18, Rab8, Rab5, Rab7 and Rab6 in animal, indicating that Rab family has further differentiation in plant (Pereira-Leal and Seabra 2001; Brighouse et al. 2010).
Rab protein family members have about 200 amino acids. Their sequences are conservative with high sequence similarity. All Rab proteins have five typical conserved domains, including four guanine nucleotide binding domains (G1, G3, G4 and G5) and an effector binding domain (G2) (Takai et al. 2001; Agarwal et al. 2009). Four domains of G1, G3, G4 and G5 participate in the binding and hydrolysis of nucleotides. Among them, G1 is the binding site of phosphate or Mg2+, G4 and G5 are key sites involved in GTP-GDP binding and hydrolysis (Stenmark and Olkkonen 2001). Mutations in the important amino acid sites of these conserved domains will cause Rab proteins to produce some constitutive inhibiting or activating mutant proteins. Normally, the amino acid sequence at the C end of the Rab protein is highly variable, but it ends with two conserved cysteine residues (CC) ultimately. These two highly conserved cysteine residues play an important role in membrane localization and protein function (Rutherford and Moore 2002). Although five G domains of Rab protein is discrete distribution in amino acid sequence, they are close to each other to form a special catalytic domain in the three-dimensional conformation of proteins, so as to better exercise the function of a protein (Rutherford and Moore 2002). The main function of the Rab protein family member is responsible for intracellular protein transport and they are essential regulator of vesicle trafficking way (Novick and Zerial 1997; Brennwald 2000). Newly synthesized secretory proteins are usually transported from one compartment of the organelle to another membrane through vesicles (Gurkan et al. 2005). They are transported to the endoplasmic reticulum at first, then transported to the plasma membrane through the Golgi apparatus, and some are delivered to the lysosome. In general, Rab proteins were involved in various cellular physiological functions of vesicular transport, such as cell polarity, cytokinesis, cell plate formation and so on (Barr 2009). Therefore, vesicular transport includes four steps at least. Vesicles budded from the donor membrane, moved to the receptor, anchored in the membrane receptor, and fused with membrane receptors. These processes all need the active involvement of Rab proteins (Tuvim et al. 2001; Yang 2002).
Up to now, the identification of Rab protein gene family in Arabidopsis (Rutherford and Moore 2002) has been very clear. But the related gene family research has not been reported in cotton. As is known to all, Rab gene family is a vital family in plant (Hill and Sylvester 2007). As one of the most important families in cotton, the Rab gene family plays a key role in the process of fiber development and biotic and abiotic stresses. With the great progress of genome sequence information in four different cotton species (G. raimondii, G. arboreum, G. hirsutum acc. TM-1 and G. barbadense acc. 3-79) (Wang et al. 2012; Li et al. 2014; Zhang et al. 2015; Yuan et al. 2015), we tried to mine the important genes related to fiber development and biotic and abiotic stresses in the whole genomic level for their functional analysis.
In this study, combined with the released genome information in four sequenced cotton species, the cotton Rab gene family members were systematically studied. We analyzed the characteristics on the gene structures, classification, chromosomal locations, and expression patterns of Rab gene family members. Our studies will lay a foundation for understanding the functional roles of different Rab members in the polar growth and stress tolerance in cotton.
Methods
Identification on Rab genes
Using the known Rab amino acid sequences (Accessions NP_187823.1 and NP_199607.1), we searched the seed file of Rab structure in the Pfam database (http://www.phytozome.net/) website. The number of seed file is PF00071. After that, the seed file was used to further retrieve the whole genome database of G. raimondii (Paterson et al. 2012) by HMMER 3.0 software (Finn et al. 2011). Finally, all obtained Rab proteins were used the blastp program on the NCBI site to further validate the conserved domains of proteins. The gene information of the other three cotton species, G. arboreum, G. hirsutum acc. TM-1, and G. barbadense acc. 3-79, were downloaded from http://cgp.genomics.org.cn, http://mascotton.njau.edu.cn and http://cotton.cropdb.org, respectively.
Chromosomal mapping and gene duplication
We used Mapchart 2.2 software to anchor the Rab genes to all 13 chromosomes of G. raimondii. The syntenic information of G. raimondii was downloaded from the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication/). Rabs were mapped to the syntenic blocks for intra- and inter-genomic comparison (Xu et al. 2016a, b). The chromosome numbers were integrated with the interspecific genetic map (D1 to D13) in allotetraploid cultivated cotton species (Wang et al. 2015) and the scaffolds (Chr.1 to Chr.13) in the genomic data of G. raimondii (Paterson et al. 2012).
The timing of segmental duplication events can be estimated by computing mean Ks values for all anchor points located in the corresponding syntenic block (Paterson et al. 2012; Wang et al. 2012), and all the Ks values were parsed from PGDD syntenic data. Genes separated by five or fewer genes within a 100-kb region on a chromosome may have resulted from tandem duplication (Wang et al. 2010).
Calculating Ka and Ks
Coding sequences were aligned using the PRANK codon model with the default options, and alignment gaps were deleted manually (Loytynoja and Goldman 2005). On the basis of the aligned coding sequences, the non-synonymous substitutions per non-synonymous site (Ka) and the synonymous substitutions per synonymous site (Ks) of homologous gene pairs were computed by the maximum likelihood method in Codeml from the PAML package v4.7 (Yang 2007).
Basic bioinformatics analysis
We use ExPASy online program (http://web.expasy.org/compute_pi/) to predict molecular weight and isoelectric point of all the Rab genes. Then we use Softberry online website (http://www.softberry.com) for subcellular localization prediction (Zhang et al. 2016).
Phylogenetic and exon–intron structural analysis
Through the ClustalX software (http://www-igbmc.u-strasbg.fr/BioInfo/), all amino acid sequences of Rab proteins obtained were used for alignment analysis (Thompson et al. 1994). Then the results were saved with the MSF format. Later, the results were edited with the Genedoc software (http://www.psc.edu/biomed/genedoc/). We used MEGA5.0 software (http://www.megasoftware.net/index.html) to construct the phylogenetic tree of Rab proteins in cotton. Checking parameter Bootstrap was set to 1000 with NJ method for operation (Saitou and Nei 1987). We compared the cDNA of Rabs in cotton with DNA sequence and analyzed the gene structure by using online site (http://wheat.pw.usda.gov/piece/GSDraw.php).
RNA-seq data analysis
The RNA-seq data for Rabs expression profile analysis was derived from TM-1 transcriptome data of Zhang et al. (2015). The data used for spatial–temporal expression analysis mainly includes the expression data of tissues and organs at different stages of growth and development.
Using RNA-seq data in the different tissues of TM-1, we filtered out the reads of multiple loci mapping, and only gene-specific read counts with unique mapping were remained to calculate the gene FPKM value, where FPKM referred to fragments per kilobase of exon model per million mapped reads with Cufflinks software (http://cufflinks.cbcb.umd.edu/). Analysis method of data model expression as follows: if the value of log2 FPKM is more than 1, we consider it to be expressed and if the value of log2 FPKM is less than or equal to 1, we consider it not to be expressed (Xu et al. 2016a, b).
Results
Genome-wide identification of the Rab gene family in Gossypium
Using the four sequenced cotton species (G. raimondii, G. hirsutum acc. TM-1, G. barbadense acc. 3-79 and G. arboreum), genome-wide identification of the Rab gene family were carried out in Gossypium. With several known amino acid sequences of the Rab gene family members, seed file (PF00071) about the Rab domain was queried on the website of Pfam data base. Then the protein databases of Gossypium were searched using HMMER 3.0 software. At last, the Rab domain was confirmed by a blastp program, and 87 GrRabs, 169 GhRabs, 136 GbRabs and 80 GaRabs were obtained from the four cotton species, respectively (Additional file 1).
In allotetraploid cotton G. hirsutum acc. TM-1, the number of Rab genes is about twice as many as in diploid cotton G. raimondii and G. arboreum. By comparison, in allotetraploid cotton G. barbadense acc. 3-79, the number of Rab genes is much less than that of G. hirsutum acc. TM-1. This might result from different sequencing methods, assembly error in partial chromosomal regions, or different degrees of colonization during the evolutionary process of Gossypium, and need to be further confirmed. Especially in RabE group, the number of Rab genes in G. barbadense has decreased drastically, implying that Rab members have further differentiation of function in the group RabE of G. barbadense.
The basic bioinformatics of 87 Rabs in G. raimondii were analyzed by using various softwares and online tools (Table 1). We found that the number of amino acids about 87 GrRabs ranged from 200 to 258 aa, which was consistent with the previous reports that the Rab protein usually has about more than 200 amino acids. It was also reported that the molecular weight of Rab proteins is generally at 20–30 kDa. In the present study, we also found that all members follow this rule, in which the minimum Rab protein has a molecular weight of 21.77 kDa and the maximum 28.65 kDa.
The nomenclature of the Rab gene family in Gossypium
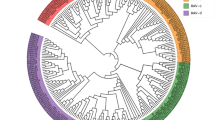
In total, 57 Rabs in A. thaliana were identified. Then they were divided into eight groups and eighteen subgroups. These groups were named as AtRabA, AtRabB, AtRabC, AtRabD, AtRabE, AtRabF, AtRabG and AtRabH, respectively. According to cluster analysis of 57 members in A. thaliana and 87 in G. raimondii, we found that 87 GrRabs were also divided into eight groups, and named as GrRabA, GrRabB, GrRabC, GrRabD, GrRabE, GrRabF, GrRabG and GrRabH, correspondingly. There were 17 subgroups in G. raimondii compared with 18 subgroups in A. thaliana (Fig. 1). The number of each subgroup was named by combining their chromosome order (D1 to D13) with their location on the chromosome. Based on the nomenclature of G. raimondii, the corresponding Rab orthologs in G. arboreum, G. hirsutum acc. TM-1, and G. barbadense acc. 3-79 were named, respectively, with the same number for orthologs as in G. raimondii (Additional file 1).
Phylogenetic relationships of Rab family members between G. raimondii and Arabidopsis. The multiple alignment of the conserved Rab family domain between G. raimondii and Arabidopsis were constructed with Clustal X (version 2.0). Phylogenetic tree was generated using the maximum likelihood method under WAG model in MEGA v5.2, and the reliability of interior branches was assessed with 1000 bootstrap resamplings
We counted the numbers and proportion of Rab genes of the eight groups between Arabidopsis and G. raimondii (Tables 2 and 3). As we can see that RabA is the largest group of the family and the largest proportion. The proportion of the same group was similar between Arabidopsis and G. raimondii. According to the position of each member in the phylogenetic tree, combining with the function of reported genes in the same group, we speculated that there were the similar function in the same group.
The chromosome distribution of the Rab gene family in G. raimondii
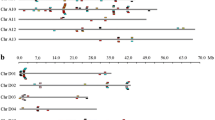
To elucidate the chromosomal distribution of these Rab genes, we integrated 13 scaffolds of the G. raimondii genome (named as Chr. 1 to Chr. 13) with the reported high-density interspecific genetic map of allotetraploid cultivated cotton species (Wang et al. 2015). Using Mapchart 2.2 software and the genomic localization information, we mapped the distribution of 87 GrRab genes on 13 chromosomes of G. raimondii (Fig. 2). 85 GrRab genes were tagged on the corresponding physical location of all the 13 chromosomes except for two genes, indicating that the scaffolds involved in the two genes had not been integrated in the chromosome in the present draft genome. The chromosomal distribution patterns of these GrRabs were uneven. Chr.11 (D10) contained the most Rabs (11 genes), while Chr.12 (D4) contained the fewest (two genes).
Chromosomal distribution of Rab genes in G. raimondii. The chromosome numbers were consistent with the interspecific genetic map (D1 to D13) in allotetraploid cultivated cotton species (Wang et al. 2015) and the scaffolds (Chr.1 to Chr.13) in the genomic data of G. raimondii (Paterson et al. 2012). The nomenclature of Rabs was based on Arabidopsis nomenclature and the order of the chromosomes in G. raimondii
Rab gene family expansion in G. raimondii
To investigate the Rab gene family expansion pattern in cotton, we download the syntenic data of G. raimondii from the Plant Genome Duplication Database (PGDD). We identified the number of Rab genes formed in whole genome duplication (WGD) and tandem duplication in cotton. Among the 87 GrRabs in G. raimondii, we identified 60 pairs of GrRabs formed in whole genome duplication (WGD) (Additional file 2). We also analyzed the adjacent genes to investigate whether tandem duplication had taken place. Four of the 87 GrRabs showed tandem duplication. They were GrRabA2a vs GrRabD1a and GrRabA2h vs GrRabD1b respectively.
In 60 pairs of whole genome duplication gene pairs (Additional file 2), we obtained the Ks values of 58 pairs of whole genome duplication gene pairs. Of them, the Ks values of eight pairs of genes are greater than 1. They were GrRabB1b vs GrRabB1a, GrRabC2c vs GrRabC2d, GrRabE1a vs GrRabE1g, GrRabE1e vs GrRabE1b, GrRabE1b vs GrRabE1f, GrRabE1b vs GrRabE1g, GrRabE1e vs GrRabE1f and GrRabE1g vs GrRabE1h, respectively. It was found that the majority of the gene pairs come from GrRabE group. These gene pairs may be derived from the ancient event. For the remaining 50 gene pairs, the Ks values ranged from 0.331 to 0.8772, implying that these gene pairs originated from the Gossypium lineage WGD events. Among all the gene pairs, the Ka/Ks values were less than 1, which indicated the results of the purification selection, and help maintain the conservation of the duplicated genes in structure and function.
Exon–intron structural analysis and subcellular localization prediction
In order to better understand the genetic structure of 87 Rab gene family members in G. raimondii, we obtained the exon and intron structure distribution of each gene by combining their genome sequence with CDS sequence (Fig. 3) using the online website (http://wheat.pw.usda.gov/piece/GSDraw.php). The exon and intron gene structure analysis showed that all 87 Rabs had different introns. It is worth mentioning that the number of introns is different, but the genes clustered together have a very similar distribution of exons and introns. For 87 Rab protein gene family members, we compared the numbers of introns in each class. In all of 34 RabA family members, we found that there is only one intron among the 33 Rabs with one exception. Among 6 RabB family members, there are 5 introns. Then, we found that there are 5 introns among the most of RabC family members. All Rabs in RabD and RabE family contain 7 introns. Among 9 RabF family members, there are 8 Rabs containing 6 introns. Finally, all Rabs contained 6 introns in RabG family and 5 introns in RabH family. It can be concluded that intron numbers of most Rabs in the same group are basically the same, only a few genes exception.
Phylogenetic relationships and gene structures of Rabs in cotton. The multiple alignment of the conserved Rab family domain were constructed with Clustal X (version 2.0). Phylogenetic tree was generated using the maximum likelihood method under WAG model in MEGA v5.2, and the reliability of interior branches was assessed with 1000 bootstrap resamplings. The gene structures were drawn using the online tool GSDS. Introns and exons were represented by black lines and blue boxes, respectively, and numbers at the exon–intron joints were intron phases
Using the Softberry online software for subcellular localization prediction, we found that 87 Rabs mainly concentrated in organelles with membrane structure and cell membrane (Table 1). We further found that all members of RabA, RabB and RabH are located in the Golgi, RabGs are located in the vacuole, RabCs are distributed in the cell membrane or Golgi, RabDs and RabFs are distributed in the cytoplasmic or Golgi, RabEs are distributed in the cell membrane or cytoplasmic.
Taken together, the genes clustered together have similar gene structure and subcellular localization, implying Rab genes in the same class generally have similar functions.
Expression pattern analysis
The analysis of gene expression patterns can provide powerful clues and help for prediction of gene function. We found that cotton Rabs are expressed in all tissues of plant and are regulated temporally and spatially depending on developmental stage and environmental conditions (Additional files 3, 4). Among the 169 GhRabs (Fig. 4), there were 161 genes with log2 FPKM > 1 in at least one of the 16 investigated organs and developmental stages, and these 161 genes were used to gauge the relative expression of each Rab gene. It indicates that most Rab family members play a certain role in different growth stages of cotton. The remaining 8 GhRabs may be pseudogenes or only expressed under special environmental conditions. About half of the genes (86 GhRabs) were expressed widely in vegetative organs, flower organs and reproductive organs. It shows that they are constitutively expressed and participate in the whole growth and development process of cotton. Approximately 10.65% (18 GhRabs) of the genes were expressed preferentially in cotton fibers, and most of them were highly expressed at the elongation stage of fiber development, indicating that these genes may be involved in regulating the growth and development of cotton fiber. A few genes are dominantly expressed in floral organs, suggesting that they may play a role in pollen formation and development. A small number of genes were preferentially expressed in the root, stem and leaf, suggesting that they might be involved in the process of plant stress resistance. We also observed the significant different expression patterns among each group of Rab family.
Expression profiles of Rab genes in cotton. The RNA-seq relative expression data of 16 tissues was used to re-construct expression patterns of 169 Rab genes in G. hirsutum acc. TM-1. Analysis method of data model expression as follows: if the value of log2 FPKM is more than 1, we consider it to be expressed and if the value of log2 FPKM is less than or equal to 1, we consider it not to be expressed
Discussion
It is reported that Rab genes played the various roles in all kinds of plants. Through cotton genome sequence information, we tried to analyze the basic biological information, gene structure, genome distribution, and expression characteristics of Rab gene family members. We hope to find some important genes related to stress response and fiber development in the whole genomic level, and help to utilize them to improve the quality and yield of cotton.
Consistency on classification and structure of 87 GrRabs
In Arabidopsis, a total of 57 Rab members are divided into eight groups. Similarly, 87 Rabs in G. raimondii are also divided into eight groups. We found that although the length of 87 Rabs and the numbers of introns are different, it is worth mentioning that the genes clustering together have a very similar distribution of exon and intron structure and subcellular localization. The intron numbers of 87 GrRabs in the same group are basically the same. It is believed that the genes clustered together have similar gene structure and the genes in the same class have similar functions.
Previous studies on many Rab proteins in Arabidopsis have been reported in detail. However, the study of Rab proteins in cotton is few. According to the function of Rab proteins in Arabidopsis (Rutherford and Moore 2002), we can infer the possible function roles of Rab proteins in cotton by clustering Rab members in Arabidopsis and G. raimondii. It has a guiding role for further study of gene function by genomic-wide comparative analysis among different plant species.
The functional diversity of Rab proteins
We used the transcriptome sequencing results of upland cotton genetic standard line TM-1 and carried out expression pattern analysis on 169 GhRabs. We found that 161 of 169 GhRabs have a certain amount of expression level in at least one of the 16 investigated organs and developmental stages. We speculate that the vast majority of Rab genes have certain function in cotton growth and development. About half of the genes (86 GhRabs) were expressed in all kinds of tissues and organs. In addition, some genes specifically expressed in certain tissues and organs, implying that Rab genes have diversity on the function. Referring the reports of Rabs function and their expression profile in cotton, cotton Rabs may involve in the biotic and abiotic stress response, pollen germination and pollen tube elongation, seed germination and fiber development.
In the previous study, it is reported that many Rab proteins are involved in important functions in a lot of plants. Among them, the hottest spot of current research is that Rab protein was involved in cell polarity growth. In the plant growth process, polar growth process is very important. The polarity growth process in plant includes elongation of pollen tubes (Peng et al. 2011) and root hairs (Preuss et al. 2006; Blanco et al. 2009). This process requires vesicle trafficking and microtubule actin proteins. Rab protein is an important molecular switch in the cell, which controls the transport of vesicles in various ways, so as to promote the establishment and maintenance of apical polarity (Cole and Fowler 2006; Samaj et al. 2006).
In Arabidopsis, root hairs and pollen tubes elongate through the polar growth (Campanoni and Blatt 2007). Although they have different origins and functions, these cells promote cell elongation by Ca2+ enrichment at the cell tip and actin cytoskeleton regulation (Li and Yang 1999; Hepler et al. 2001; Samaj et al. 2005; Cole and Fowler 2006). The overexpression of Rab11b inhibits the elongation of pollen tube and influence the direction of pollen tube growth (de Graaf et al. 2005). NtRab2, which is overexpressed in tobacco, also inhibits the growth of pollen tube (Cheung et al. 2002). In Arabidopsis, RabA4d is necessary for the proper regulation of pollen tube growth. The loss of RabA4d leads to the destruction of polar growth and changes in the structure of the cell wall. These results indicate that RabA4d plays an important role in regulating the growth of pollen tube (Szumlanski and Nielsen 2009). Enhanced yellow fluorescent protein RabA4b-EYFP specifically located in the tip of the root hair cells of Arabidopsis. In root hair defective mutants, yellow fluorescence signal localization of RabA4b-EYFP in hair cells no longer exist. As a result, RabA4b potentially regulates membrane transport in plant cells through the involvement of cell wall components in the secretory process (Preuss and Nielsen 2004). AtRab2 was specifically expressed in newly germinated seedlings and pollen tubes. The GUS connected to the promoter of the gene was clearly detected in mitosis of pollen tubes. After that, its expression level has been increased, indicating that AtRab2 plays an important role in the growth and development of pollen tubes (Moore et al. 1997). Rgp1 isolated from rice is a gene homologous to yeast Ypt3. After overexpression of this gene in tobacco plants, the plants exhibited dwarf and the abnormal development of floral organs. Of them, the transgenic plants were more than 6 times higher in the content of endogenous hormones than that in the wild plants. These results suggest that Rgp1 may be involved in cytokinin signaling pathways (Kamada et al. 1992; Sano et al. 1994). Thus, the Rab protein is essential for root hairs and pollen tube elongation of plant. Cotton fiber cells, root hairs and pollen tubes are typical tissues for studying polar growth of plants. We hypothesized that Rab proteins would be involved in the fiber polarity elongation process and plant growth process in cotton.
Rab proteins not only play an important role in plant growth and development, but also play an important role in biotic and abiotic stress responses. When the relative water content of leaves decreased, the expression of Rab2 increased. Further, the expression level of Rab2 was increased when exogenous ABA was applied to the restored grass plants. The results show that Rab2 plays an important role in drought stress (O’Mahony and Oliver 1999). Under a variety of environments (chilling, salinity, drying and ABA) induction, the expression level of OsRab7 significantly altered. Through the transformation of Arabidopsis protoplasts, the OsRab7 fusion protein of GFP was localized in the vacuole. It is suggested that OsRab7 is transported to the vacuole through the vesicle transport in plant cells. The inducible expression of OsRab7 suggests that it is involved in the response of these stresses, which suggests that Rab is related to abiotic stresses (Nahm et al. 2003). Overexpression of AtRabG3e in Arabidopsis could enhance the resistance of plants to salt and osmotic stress. The enhancement of plant resistance was related to the acceleration of endocytosis and the increase of sodium ion in vacuole (Mazel et al. 2004). In addition, when AtRabG3b was silenced, there was no difference in the silenced plants compared with the no transgenic plants. However, when AtRabG3b was overexpressed, it showed hypersensitive cell death on pathogenic fungi and mycotoxins. In addition, it also accelerated leaf senescence. These results suggest that AtRabG3b is involved in the regulation of cell apoptosis and the regulation of pathogen response during plant senescence (Kwon et al. 2009). Transgenic tobacco plants overexpressing PgRab7 also showed resistance to drought and salt stress, which indicated that PgRab7 was involved in plant stress response (Agarwal et al. 2008). OsRab11 is widely expressed in various tissues and organs of plants, which is induced by jasmonic acid (JA). Overexpression of OsRab11 in transgenic plants enhanced the resistance to pathogens by affecting the expression of genes involved in the metabolic pathway of jasmonic acid. Therefore, OsRab11 is necessary in the jasmonic acid mediated signaling pathway (Hong et al. 2013). Based on above, we hypothesized that Rab genes in cotton play an important role in biotic and abiotic stress responses.
Conclusions
Rab proteins play an important role in plant growth and development, as well as in biotic and abiotic stress responses. In this study, we individually identified 87, 169, 136, 80 Rabs in the four sequenced cotton species. These Rabs are divided into eight groups. In each group, their intron numbers and subcellular localization are basically the same. Further, 60 pairs of segmental duplication due to whole genome duplication and two pairs of tandem duplication were detected, respectively. Expression patterns analysis indicated that most Rab family members play a certain role in different tissues/organs and different growth stages of cotton, implying their potential function in the polar growth and stress tolerance.
Abbreviations
- Rab :
-
ras-like in rat brain
- PGDD:
-
plant genome duplication database
- WGD:
-
whole genome duplication
- FPKM:
-
fragments per kilobase of exon model per million mapped reads
- Ka :
-
nonsynonymous substitutions per nonsynonymous site
- Ks :
-
synonymous substitutions per synonymous site
References
Agarwal PK, Agarwal P, Jain P, Jha B, Reddy MK, Sopory SK (2008) Constitutive overexpression of a stress-inducible small GTP-binding protein Pgrab7 from Pennisetum glaucum enhances abiotic stress tolerance in transgenic tobacco. Plant Cell Rep 27:105–115
Agarwal P, Reddy MK, Sopory SK, Agarwal PK (2009) Plant Rabs: characterization, functional diversity, and role in stress tolerance. Plant Mol Biol Rep 27:417–430
Barr FA (2009) Rab GTPase function in Golgi trafficking. Semin Cell Dev Biol 20:780–783
Blanco FA, Peltzer Meschini E, Zanetti ME, Aguilar OM (2009) A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21:2797–2810
Brennwald P (2000) Reversal of fortune: do Rab GTPases act on the target membrane? J Cell Biol 149:1–4
Brighouse A, Dacks JB, Field MC (2010) Rab protein evolution and the history of the eukaryotic endomembrane system. Cell Mol Life Sci 67:3449–3465
Campanoni P, Blatt MR (2007) Membrane trafficking and polar growth in root hairs and pollen tubes. J Exp Bot 58:65–74
Cheung AY, Christine Y-HC, Glaven RH, Graaf BHJD, Luis V, Hepler PK, Wu HM (2002) Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14:945–962
Cole RA, Fowler JE (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9:579–588
de Graaf BH, Cheung AY, Andreyeva T, Levasseur K, Kieliszewski M, Wu HM (2005) Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell 17:2564–2579
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:29–37
Gallwitz D, Donath C, Sande C (1983) A yeast gene encoding a protein homologous to the human c-has/bas proto-oncogene product. Nature 306:704–707
Gurkan C, Lapp H, Alory C, Su AI, Hogenesch JB, Balch WE (2005) Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell 16:3847
Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17:159–187
Hill D, Sylvester A (2007) Diversification of the Rab guanosine triphosphatase family in dicots and monocots. J Integr Plant Biol 49:1129–1141
Hong MJ, Yun ML, Son YS, Im CH, Yi YB, Rim YG, Bahk JD, Heo JB (2013) Rice Rab11 is required for JA-mediated defense signaling. Biochem Biophys Res Commun 434:797–802
Kamada I, Yamauchi S, Youssefian S, Sano H (1992) Transgenic tobacco plants expressing rgpl, a gene encoding ras-related GTPbinding protein from rice, show distinct morphological characterization. Plant J 2:799–807
Kwon SI, Hong JC, Bae K, Jung JH, Jin HC, Park OK (2009) Role of an Arabidopsis, Rab GTPase RabG3b in pathogen response and leaf senescence. J Plant Biol 52:79–87
Li H, Yang Z (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11:1731–1742
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2014) Genome sequence of the cultivated cotton Gossypium arboretum. Nat Genet 46:567–572
Loytynoja A, Goldman N (2005) An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA 102:10557–10562
Martinez O, Goud B (1998) Rab proteins. Biochim Biophys Acta 1404:101–112
Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134:118–128
Moore I, Diefenthal T, Zarsky V, Schell J, Palme K (1997) A homolog of the mammalian GTPase Rab2 is present in Arabidopsis and is expressed predominantly in pollen grains and seedlings. Proc Natl Acad Sci USA 94:762–767
Nahm MY, Kim SW, Yun D, Lee SY, Cho MJ, Bahk JD (2003) Molecular and biochemical analyses of OsRab7, a rice Rab7 homolog. Plant Cell Physiol 44:1341–1349
Novick P, Zerial M (1997) The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol 9:496–504
O’Mahony PJ, Oliver MJ (1999) Characterization of adesiccation-responsive small GTP-binding protein (Rab2) from thedesiccation-tolerant grass Sporobolus stapfianus. Plant Mol Biol 39:809–821
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Peng J, Ilarslan H, Wurtele ES, Bassham DC (2011) AtRabD2b and AtRabD2c have overlapping functions in pollen development and pollen tube growth. BMC Plant Biol 11:1–16
Pereira-Leal JB, Seabra MC (2001) Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313:889–901
Preuss ML, Nielsen E (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16:1589–1603
Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E (2006) A role for the RabA4b effector protein PI-4K1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol 172:991–998
Rutherford S, Moore I (2002) The Arabidopsis, Rab GTPase family: another enigma variation. Curr Opin Plant Biol 5:518–528
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salminen A, Novick PJ (1987) A ras-like protein is required for a post-Golgi event in yeast secretion. Cell 49:527–538
Samaj J, Read ND, Volkmann D, Menzel D, Baluska F (2005) The endocytic network in plants. Trends Cell Biol 15:425–433
Samaj J, Muller J, Beck M, Böhm N, Menzel D (2006) Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci 11:594–600
Sano H, Seo S, Orudgev E, Youssefian S, Ishizuka K (1994) Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response towounding, and increases resistance to tobacco mosaic virus infection. Proc Natl Acad Sci USA 91:10556–10560
Stenmark H, Olkkonen VM (2001) The Rab GTPase family. Genome Biol 2(reviews3007):1
Szumlanski AL, Nielsen E (2009) The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21:526–544
Takai Y, Sasaki T, Matozaki T (2001) Small GTP-binding proteins. Physiol Rev 81:153–208
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W, (improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice). Nucleic Acids Res 22:4673–4680
Tuvim MJ, Adachi R, Hoffenberg S, Dickey BF (2001) Traffic control: Rab GTPases and the regulation of interorganellar transport. News Physiol Sci 16(2):56–61
Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L (2010) Expression profiling and integrative analysis of the CESA/CSL, superfamily in rice. BMC Plant Biol 10(1):282
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44(10):1098–1103
Wang S, Chen JD, Zhang WP, Hu Y, Chang LJ, Fang L, Wang Q, Lv FN, Wu HT, Si ZF, Chen SQ, Cai CP, Zhu XF, Zhou BL, Guo WZ, Zhang TZ (2015) Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol 16:108
Xu J, Xu X, Tian L, Wang G, Zhang X, Wang X, Guo W (2016a) Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci Rep 6:29022
Xu X, Feng Y, Fang S, Xu J, Wang X, Guo W (2016b) Genome-wide characterization of the β-1,3-glucanase gene family in Gossypium by comparative analysis. Sci Rep 6:29044
Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell 14:S375–S388
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, Chen L, He Y, Zhang L, Zhu L, Li Y, Liang Q, Lin Z, Yang X, Liu N, Jin S, Lei Y, Ding Y, Li G, Ruan X, Ruan Y, Zhang X (2015) The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep 5:17662
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537
Zhang X, Xu X, Yu Y, Chen C, Wang J, Cai C, Guo W (2016) Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules in cotton. Sci Rep 6:29781
Authors’ contributions
GWZ conceived and designed the experiments. LP performed the experiments, analyzed the data and wrote the paper. Both authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data and material obtained in this paper are available.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Funding
This program was financially supported in part by the JCIC-MCP project (No. 10).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Li, P., Guo, W. Genome-wide characterization of the Rab gene family in Gossypium by comparative analysis. Bot Stud 58, 26 (2017). https://doi.org/10.1186/s40529-017-0181-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40529-017-0181-y