Abstract
Background
The incidence of oropharyngeal squamous cell carcinoma (OPSCC) caused by oncogenic human papillomavirus (HPV) is rising worldwide. HPV-OPSCC is commonly diagnosed by RT-qPCR of HPV E6 and E7 oncoproteins or by p16 immunohistochemistry (IHC). Droplet digital PCR (ddPCR) has been recently reported as an ultra-sensitive and highly precise method of nucleic acid quantification for biomarker analysis. To validate the use of a minimally invasive assay for detection of oncogenic HPV based on oropharyngeal swabs using ddPCR. Secondary objectives were to compare the accuracy of ddPCR swabs to fresh tissue p16 IHC and RT-qPCR, and to compare the cost of ddPCR with p16 IHC.
Methods
We prospectively included patients with p16+ oral cavity/oropharyngeal cancer (OC/OPSCC), and two control groups: p16− OC/OPSCC patients, and healthy controls undergoing tonsillectomy. All underwent an oropharyngeal swab with ddPCR for quantitative detection of E6 and E7 mRNA. Surgical specimens had p16 IHC performed. Agreement between ddPCR and p16 IHC was determined for patients with p16 positive and negative OC/OPSCC as well as for healthy control patients. The sensitivity and specificity of ddPCR of oropharyngeal swabs were calculated against p16 IHC for OPSCC.
Results
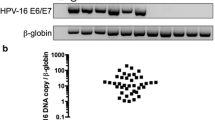
122 patients were included: 36 patients with p16+OPSCC, 16 patients with p16−OPSCC, 4 patients with p16+OCSCC, 41 patients with p16−OCSCC, and 25 healthy controls. The sensitivity and specificity of ddPCR of oropharyngeal swabs against p16 IHC were 92 and 98% respectively, using 20–50 times less RNA than that required for conventional RT-qPCR. Overall agreement between ddPCR of tissue swabs and p16 of tumor tissue was high at ĸ = 0.826 [0.662-0.989].
Conclusion
Oropharyngeal swabs analyzed by ddPCR is a quantitative, rapid, and effective method for minimally invasive oncogenic HPV detection. This assay represents the most sensitive and accurate mode of HPV detection in OPSCC without a tissue biopsy in the available literature.
Similar content being viewed by others
Background
Head and neck squamous cell carcinoma (HNSCC) is now the fifth most common malignancy worldwide [1]. The incidence of oropharyngeal squamous cell carcinoma (OPSCC), a subsite of HNSCC, is rapidly increasing in North America [2]. Although traditional risk factors for HNSCC include smoking and alcohol use, the rising incidence of OPSCC has been attributed largely to the Human Papillomavirus (HPV) [3–6]. Determining HPV positivity is of critical importance in the diagnosis and management of OPSCC, as HPV positive tumours have unique pathologic and clinical characteristics that have implications for prognosis and treatment decisions [3, 7–10].
The gold standard for determining HPV status in OPSCC is demonstration of oncogenic HPV DNA in fresh tissue using real-time quantitative polymerase chain reaction (RT-qPCR) [1, 11, 12]. Due to the high cost and specialized equipment required for this method, most centers have adopted p16 immunohistochemistry (p16 IHC) as the preferred method of oncogenic HPV detection, which has become the clinical standard. [13–17] However, p16 IHC is an imperfect surrogate marker for HPV-associated OPSCC. HPV infection is carcinogenic through expression of oncogenic proteins E6 and E7 which cause multiple genetic and metabolic effects within the cell, the most important of which is degradation of tumor suppressor genes including p53 and Rb [18]. By a separate pathway, this results in downstream overexpression of p16. Because p16 overexpression can occur through HPV-independent pathways, p16 IHC can lead to false positives, which could result in erroneous de-intensification of treatment. Although RT-PCR of E6 and E7 can circumvent the limitations of p16 IHC, it also requires adequate nucleic acid sample generally only attainable from tissue biopsy.
Droplet digital polymerase chain reaction (ddPCR) is a relatively novel technique with potential applications for HPV detection in OPSCC. ddPCR is the most accurate and sensitive mode of measuring target nucleic acids quantitatively in the available literature [19]. ddPCR involves partitioning a single nucleic acid sample in up to 20,000 discrete water-in-oil droplets and performing PCR analysis on each droplet independently, with the results reported digitally and quantitatively. This technique quantifies the absolute amount of target nucleic acid present with greater precision and reproducibility than RT-qPCR [20–23]. ddPCR has been used to quantify gene expression with extremely low copy number [24, 25], and has demonstrated better diagnostic performance in biomarker analysis than other molecular techniques. [22, 26, 27] One study made use of ddPCR for detection of oncogenic E6/E7 mRNA in fresh tissue OPSCC specimens and found 100% sensitivity compared to p16 IHC using target RNA 20–50 times lower than reported for RT-qPCR [28].
Because of the accuracy and ultra-sensitivity of ddPCR for identification of oncogenic HPV E6/E7 mRNA, we hypothesized that ddPCR can be used for detection of oncogenic HPV in OPSCC using oral/oropharyngeal swabs as opposed to fresh tissue. The ability to detect oncogenic HPV without a tissue biopsy would have important implications for diagnosis, post-treatment surveillance, and screening of patients with OPSCC.
The purpose of this study was to validate the use of a novel minimally-invasive assay for detection of oncogenic HPV based on oral/oropharyngeal swabs using ddPCR. Our secondary objectives were to compare the accuracy of ddPCR swabs to fresh tissue p16 IHC, and to report the cost of ddPCR.
Methods
This was a single-center prospective cohort validation study at a tertiary care Otolaryngology-Head and Neck Surgery referral center in Edmonton, AB, Canada. Health research ethics board approval was obtained from the University of Alberta prior to commencement of the study (Pro00057994).
Participants
Participants were recruited at initial presentation to the University of Alberta Otolaryngology-Head and Neck Surgery Clinic between February 2015 and March 2016. Adult patients with biopsy-confirmed oral cavity or oropharyngeal squamous cell carcinoma (OC/OPSCC) were identified. Patients who had previous treatment for HNSCC, those that were unable to undergo an oropharyngeal swab, patients for which pathology report or p16 IHC was unavailable, patients with unknown primary tumours, and improperly processed samples were excluded. The reason for including OCSCC patients in addition to OPSCC was to be able to compare ddPCR and p16 in a subgroup of patients for which p16 is an especially poor marker of HPV infection, in order to determine the enhanced specificity of ddPCR for HPV oncogenesis.
The control group was recruited from patients who were being consented for tonsillectomy for a benign indication (ex: recurrent tonsillitis or obstructive sleep apnea). Patients were excluded from the control group if they had a previous history of HNSCC.
Oral/oropharyngeal swabs
Each participant underwent an oral/oropharyngeal swab using a 10 cm cotton-tip applicator by a staff Otolaryngologist (VB, HS, JH, or DO). Two swabs were performed on each patient. For patients with a clinically evident oral/oropharyngeal tumour, one swab was taken from the tumour, and the second swab was taken from the oropharynx (a single swab that was brushed against the tonsils, base of tongue, soft palate, and posterior pharyngeal wall subsites). For control patients, two swabs were taken both from the oropharyngeal subsites listed above (each swab was brushed against all of the subsites listed above). The swab tips were immediately placed in 3 mL of RNAlater (Ambion-Thermo Fisher Scientific, Waltham, MA, USA) and stored at room temperature (RT) for up to 24 h, then at 4 °C for up to 7 days prior to RNA extraction.
Tissue pathology and p16 IHC
Each patient with an oral/oropharyngeal tumour underwent pan-endoscopy with biopsy of the tumour as per standard clinical practice. Pathology was reported by a head and neck pathologist at the University of Alberta to confirm the diagnosis of SCC. p16 IHC was performed on representative 4 μm sections cut from formalin-fixed, paraffin-embedded tissue blocks using a monoclonal antibody to p16, as per established guidelines [29]. For control patients, tonsil specimens were sent for pathologic analysis at the time of tonsillectomy and were interpreted by an anatomic pathologist at the University of Alberta to confirm the diagnosis of non-malignant tonsil tissue. P16 was not performed if the tissue was deemed benign, since the significance of p16 in the absence of carcinoma is unclear. Instead, these patients were used as negative controls.
RNA extractions and cDNA synthesis
RNA was extracted from tumor tissue using the RNeasy Mini Kit (Qiagen, Germantown, MD, USA) following the manufacturer’s protocol. Salivary swab samples were vortexed in 15 mL conical tubes containing 3 mL of RNAlater and centrifuged at 3000 x g for 10 min. The RNAlater was then aspirated and cell pellet was re-suspended in 350 μL of buffer RLT containing 40 mM DTT. RNA was eluted from the mini column with 35 μL of RNAse free water. RNA concentration was quantified using the Qubit RNA HS assay kit on a Qubit 2.0 fluorometer.
Extracted RNA (100–200 ng) was used to synthesize cDNA using the iScriptTM Reverse Transcription Supermix for RT-qPCR (BIO-RAD) as per the manufacturer’s protocol. Following the reaction the cDNA was diluted with 0.125 mM EDTA pH 8.0 to 0.5 ng/μl and either stored at −20 °C or used directly for ddPCR.
Droplet digital PCR
All ddPCR reactions were performed by MK, who was blinded to the patient group, pathology, and p16 IHC status of participant samples. ddPCR was carried out using the ddPCRTM Supermix for Probes (No dUTP) (BIO-RAD, Mississauga, ON, CAN), the QX200TM Droplet Generator (catalog #186-4002 BIO-RAD), the QX200 Droplet Reader (catalog #186-4003 BIO-RAD) the C1000 TouchTM Thermal Cycler (catalog #185-1197 BIO-RAD) and the PX1TM PCR Plate Sealer (catalog #181-4000 BIO-RAD) as per manufacturer’s instructions. Reactions were set up following the manufacturer’s protocols using 12 μl/reaction of 2x ddPCR Supermix for Probes (No dUTP), 1.2 μL/reaction of 20x target primers/probe (FAM or HEX, BIO-RAD), 1.2 μL/reaction 20x reference primers/probe (FAM or HEX, BIO-RAD), 2.4 μL cDNA (at 0.5 ng/μl) and 7.2 μl H2O. Human EEF2 primers/probes (BIO-RAD) were used as an internal reference standard and indirect indicator of nucleic acid stability in participant samples. HPV E6 and E7 ddPCR detection was performed using the following primer/probe sequences generated by BIO-RAD [30], adapting primer sequences from HPV E6: forward sequence, 5′-TCAGGACCCACAGGAGCG-3′, reverse sequence, 5′-CCTCACGTCGCAGTAACTGTTG-3′, probe (FAM-labeled) sequence, 5′-CAGAAAGTTACCACAGTTATGCACAGAGCT-3′. HPV E7: forward sequence, 5′-CCGGACAGAGCCCATTACAA -3′, reverse sequence, 5′-CGAATGTCTACGTGTGTGCTTTG -3′, probe (HEX-labeled) sequence, 5′-CGCACAACCGAAGCGTAGAGTCACACT -3′. Reactions were set up in a 96 well plate, mixed using a Mixmate Vortex Shaker (Eppendorf, Mississauga, ON, CAN) and 20 μl of the reaction mixture was transferred to DG8TM Cartridge for QX200/QX100 Droplet Generator (catalog #186-4008 BIO-RAD) followed by 70 μl of Droplet Generation Oil for Probes (catalog #186-3005 BIO-RAD) into the oil wells, according to the QX200 Droplet Generator Instruction Manual (#10031907 BIO-RAD). Following droplet generation, 40 μL of the reaction was transferred to wells of a 96 well plate. The plates were sealed and the reactions were carried out in the thermocycler using the following parameters: Step 1) 95 °C for 10 min, Step 2) 94 °C for 30 s and 60 °C for 1 min (Step 2 repeat 39 times for a total of 40), Step 3) 98 °C for 10 min and Step 4) 4 °C infinite hold. All steps had a ramp rate of 3 °C/s. Following thermocycling the reactions were read in the QX200 Droplet Reader and the RNA targets were quantified using the QuantaSoftTM Software (BIO-RAD). HPV E6 and E7 positivity was determined using automated cutoff values relative to control as previously described [28]. Samples with > 2 droplets in the positive range for E6 or E7 were considered positive. Samples with < 20 positive droplets were re-analyzed to ensure these low copy number samples were not due to cross-contamination.
Validation of ddPCR sensitivity against RT-qPCR
As previously shown in other studies, we confirmed the enhanced sensitivity of ddPCR over RT-qPCR using decreasing concentrations of target RNA for EEF2 and E7 (see Additional file 1: Table S1). As recommended by the manufacturer (BioRAD), the same EEF2 and E7 primer/probe sets used in ddPCR were used for qRT-PCR with optimized annealing temperatures. RT-qPCR was performed as follows: RNA (100 ng) was used to synthesize cDNA using the iScriptTM Reverse Transcription Supermix for RT-qPCR (BIO-RAD) as per the manufacturer’s protocol in a 20 ul reaction. The resulting cDNA was diluted to 1.25 ng/ul, 0.125 ng/ul and 0.0125 ng/ul with water and these dilutions were used to load 10 ng, 1.0 ng and 0.1 ng respectively into reaction wells for either the qPCR or ddPCR. 20 ul final qPCR reactions were set up following the manufacturer’s protocols using 10 ul/reaction of 2x iTaqTM Universal Probes Supermix (BIO-RAD), 1 ul/reaction of 20x target primers/probe (FAM or HEX, BIO-RAD), 1 ul/reaction 20x reference primers/probe (FAM or HEX, BIO-RAD) and 8 ul of cDNA. Reactions were carried out in the CFX96 Touch™ Real-Time PCR Detection System using the “Prime PCR” Manufacturer’s program with the following parameters: Step 1) 95 ° C for 2 min, Step 2) 95 ° C for 5 s and 60 ° C for 30 s (Step 2 repeat 39 times for a total of 40), Step 3) 95 ° C for 5 s.
Data analysis
Descriptive statistics were performed to determine the proportion of patients recruited with p16 positive vs. negative OC/OPSCC. Agreement between ddPCR and p16 IHC was determined for patients with p16 positive and negative OC/OPSCC as well as for healthy control patients. The sensitivity and specificity of ddPCR of oropharyngeal swabs were calculated against p16 IHC for OPSCC.
Results
122 patients met the inclusion and exclusion criteria and were prospectively enrolled in the study. These patients are summarized in Table 1. There were 36 patients with p16 positive OPSCC (p16+OPSCC), 16 patients with p16 negative OPSCC (p16−OPSCC), 4 patients with p16+OCSCC, and 41 patients with p16−OCSCC. 25 patients were healthy controls. The mean concentration of RNA obtained from oral/oropharyngeal swabs was 5.31 μg/mL (range 2.2-12.1 μg/mL). The minimum target RNA required per reaction was ≤1 ng.
33/36 (92%) of patients with p16+OPSCC tested positive for E6/E7 by ddPCR (Table 2). One patient with p16−OPSCC tested positive for E6/E7 by ddPCR. The sensitivity of ddPCR of oropharyngeal swabs against fresh tissue p16 IHC was 92%. All 4 patients with p16+OCSCC tested negative for E6/E7 by ddPCR, as did all 41 patients with p16−OCSCC (Table 3). All 25 healthy control patients tested negative for E6/E7 by ddPCR, for an overall specificity of 98% for OPSCC. The agreement between ddPCR and p16 via unweighted kohen’s kappa was ĸ = 0.826 [0.662-0.989]
The cost of p16 IHC across the province of Alberta is $31.10/slide, with a minimum of 2 slides required per patient (≥ $62.10/patient). In comparison, the total cost of HPV E6/E7 ddPCR including technical labour was estimated to be $20.45 per patient sample.
Discussion
The incidence of HPV-related OPSCC is rapidly growing and poses challenges for diagnosis and management. Accurate determination of HPV status in patients with OPSCC is therefore critical. Performing HPV testing using a method that is cost effective and minimally invasive while maintaining adequate sensitivity has many potential applications. The method presented here is to our knowledge the most sensitive method of diagnosing oncogenic HPV mRNA without a tissue biopsy reported to date. Every swab performed in this study yielded adequate RNA for amplification, with 1 ng of RNA/reaction required for robust ddPCR results. This is an order of magnitude less than the 20–50 ng/reaction of RNA that is normally required for RT-qPCR [30–33]. This is also to our knowledge the first study to compare non-invasive oncogenic HPV detection with the clinical reference standard of p16 IHC. In addition to the greater specificity of using an HPV-specific assay, our cost analysis demonstrates a significant cost-savings of ddPCR over p16 IHC for determining HPV status in OPSCC.
Overall our results demonstrated excellent sensitivity in detecting oncogenic HPV in oropharyngeal swabs without a biopsy, as 92% of p16+OPSCC tested positive by ddPCR. The three patients with p16+OPSCC that tested negative for E6/E7 by ddPCR may in fact highlight the limitations of p16 IHC as a surrogate marker for HPV, as these three patients were older and had significant smoking and alcohol use histories, and thus may have had disease that was unrelated to HPV. While studies have suggested that p16 is itself an important prognostic marker independent of HPV infection in OPSCC [13, 34], others have questioned the prognostic utility of p16 in HPV negative tumors [35], and have demonstrated improved prognostication when HPV-specific tests are used [36]. Robinson et al. argued that HPV-specific testing remains essential in OPSCC regardless of p16 status [37].
Our results also demonstrated a high degree of accuracy in determining HPV status in OCSCC, as all 4 patients with p16+OCSCC tested negative for E6/E7 by ddPCR. Other studies have shown that non-OPSCC that are p16 positive are usually unrelated to HPV, and that p16 positivity does not purport a better prognosis in non-OPSCC, and may in fact yield a worse prognosis in these patients [38–41]. This adds to the clinical utility of using HPV-specific ddPCR as opposed to or in addition to p16 IHC in both OPSCC and non-OPSCC.
The fact that the assay we have described yielded excellent sensitivity without the need for a tissue biopsy has several important implications, the most immediately relevant of which may be for post-treatment surveillance. One recent study claimed to be able to predict recurrence of OPSCC earlier using salivary rinses for detection of oncogenic HPV [42], however this assay was limited by a lack of sensitivity due to the large amount of RNA required for RT-qPCR. A similarly designed study by Chuang et al. reported a sensitivity of 50% in predicting recurrent OPSCC using oral rinses for HPV DNA using RT-qPCR, although this study only included 4 patients with recurrent OPSCC [43]. Ahn et al. also used oral rinses for HPV DNA to predict recurrent OPSCC and found that when combined with plasma RT-qPCR, a sensitivity of almost 70% could be achieved; however, the pre-treatment sensitivity of oral rinses was only 53% [30]. With the significantly improved sensitivity of the novel assay we have described, post-treatment surveillance may be more effective using regular oropharyngeal swabs to detect E6/E7 via ddPCR. The use of ddPCR for post-treatment surveillance needs further research however before this can be recommended or widely utilized.
Other studies that have attempted to determine HPV status using oral rinses and various PCR-based assays in a diagnostic fashion as opposed to post-treatment surveillance have reported similarly low sensitivities. Both Zhao et al. and Nordfors et al. compared RT-qPCR in oral rinses with tissue biopsies in patients with OPSCC and reported sensitivities of 30 and 68% respectively for HPV 16 DNA [44, 45]. With the improved sensitivity provided by our ddPCR-based assay, clinicians may be able to use the information diagnostically to make a diagnosis of HPV-related OPSCC sooner. This is particularly useful in settings where a biopsy may be difficult to obtain, or as part of the work-up for an unknown primary tumor. Moreover, with the rising prevalence of HPV-related OPSCC, our ddPCR assay may be a viable mode of screening high-risk patient groups for early detection or prevention of OPSCC in the future. Fahkry et al. attempted to validate a “cervical pap smear-equivalent” test for early detection of HPV-related OPSCC using a combination of oral rinses and tonsillar brush biopsies via RT-qPCR; however, they concluded that it was not feasible due to the lack of correlation between HPV DNA detection and cellular atypia [46]. The authors postulated that this was due to the difficulty in detecting HPV DNA that may be replicating deep within the tonsillar crypt epithelium, a problem that could potentially be solved by an ultra-sensitive ddPCR-based assay.
This study had limitations, which included a single center experience and a relatively small sample size especially for p16+OCSCC and p16−OPSCC. Our sensitivity and specificity data also need to be interpreted cautiously, as these were calculated against the results of p16 IHC, which is itself known to be a surrogate marker; however, our aim was to demonstrate the sensitivity of a swab-based assay against the mostly widely-used tissue-based assay for determining HPV status. We also only tested for HPV 16 as in other studies; however, HPV 16 is known to cause more than 95% of HPV-related OPSCC [46]. We plan to expand our assay to test for HPV 18 in addition to HPV 16.
Conclusion
Oropharyngeal swabs analyzed by ddPCR is a quantitative, rapid, and cost-effective method for minimally invasive oncogenic HPV detection. This assay represents the most sensitive and accurate mode of HPV detection in OPSCC without a tissue biopsy in the available literature, and has several potential applications for both diagnosis and disease surveillance.
Abbreviations
- ddPCR:
-
Droplet digital polymerase chain reaction
- HNSCC:
-
Head and neck squamous cell carcinoma
- HPV:
-
Human papillomavirus
- IHC:
-
Immunohistochemistry
- OCSCC:
-
Oral cavity squamous cell carcinoma
- OPSCC:
-
Oropharyngeal squamous cell carcinoma
- RT-qPCR:
-
Real time quantitative polymerase chain reaction
References
Hayes DN, Van Waes C, Seiwert TY. Genetic landscape of human papillomavirus-associated head and neck cancer and comparison to tobacco-related tumors. J Clin Oncol. 2015;33(29):3227–34. doi:10.1200/JCO.2015.62.1086.
Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–301. doi:10.1200/JCO.2011.36.4596.
Lowy DR, Munger K. Prognostic implications of HPV in oropharyngeal cancer. N Engl J Med. 2010;363(1):82–4. doi:10.1056/NEJMe1003607.
Marklund L, Hammarstedt L. Impact of HPV in oropharyngeal cancer. J Oncol. 2011. doi:10.1155/2011/509036.
Osazuwa-Peters N. Human papillomavirus (HPV), HPV-associated oropharyngeal cancer, and HPV vaccine in the United States-Do we need a broader vaccine policy? Vaccine. 2013;31:5500–5. doi:10.1016/j.vaccine.2013.09.031.
Shaw R, Robinson M. The increasing clinical relevance of human papillomavirus type 16 (HPV-16) infection in oropharyngeal cancer. Br J Oral Maxillofac Surg. 2011;49(6):423–9. doi:10.1016/j.bjoms.2010.06.023.
Biron VL, Mohamed A, Hendzel MJ, Underhill DA, Seikaly H. Epigenetic differences between human papillomavirus-positive and -negative oropharyngeal squamous cell carcinomas. J Otolaryngol - Head Neck Surg. 2012;41(SUPPL. 1). doi:10.2310/7070.2011.110087.
Seikaly H, Biron VL, Zhang H, et al. Role of primary surgery in the treatment of advanced oropharyngeal cancer. Head Neck. 2015. doi:10.1002/hed.24042.
Xu CC, Biron VL, Puttagunta L, Seikaly H. HPV Status And Second Primary Tumours In Oropharyngeal Squamous Cell Carcinoma. J Otolaryngol - Head Neck Surg. 2013;42. Doi:10.1186/1916-0216-42-36.
Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi:10.1056/NEJMoa0912217.
Nishat R, Behura SS, Ramachandra S, Kumar H, Bandyopadhyay A. Human papilloma virus (HPV) induced head & neck squamous cell carcinoma: a comprehensive retrospect. J Clin Diagn Res. 2015;9(6):ZE01–4. doi:10.7860/JCDR/2015/13948.6056.
Walline HM, Komarck C, McHugh JB, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and oral cavity cancers. JAMA Otolaryngol Neck Surg. 2013;139(12):1320. doi:10.1001/jamaoto.2013.5460.
Lewis JS, Thorstad WL, Chernock RD, et al. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34(8):1088–96. doi:10.1097/PAS.0b013e3181e84652.
Thomas J, Primeaux T. Is p16 immunohistochemistry a more cost-effective method for identification of human papilloma virus-associated head and neck squamous cell carcinoma? Ann Diagn Pathol. 2012;16(2):91–9. doi:10.1016/j.anndiagpath.2011.09.002.
Oguejiofor KK, Hall JS, Mani N, et al. The prognostic significance of the biomarker p16 in oropharyngeal squamous cell carcinoma. Clin Oncol (R Coll Radiol). 2013;25(11):630–8. doi:10.1016/j.clon.2013.07.003.
Lewis JS. p16 immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2012;6 Suppl 1:75–82. doi:10.1007/s12105-012-0369-0.
El-Naggar AK, Westra WH. p16 expression as a surrogate marker for HPV-related oropharyngeal carcinoma: a guide for interpretative relevance and consistency. Head Neck. 2012;34(4):459–61. doi:10.1002/hed.21974.
Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10(8):550–60. doi:10.1038/nrc2886.
Albano F, Zagaria A, Anelli L, et al. Absolute quantification of the pretreatment PML-RARA transcript defines the relapse risk in acute promyelocytic leukemia. Oncotarget. 2015;6(15):13269–77. http://www.scopus.com/inward/record.url?eid=2-s2.0-84931038270&partnerID=tZOtx3y1.
McDermott GP, Do D, Litterst CM, et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem. 2013;85(23):11619–27. doi:10.1021/ac403061n.
Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–5. doi:10.1038/nmeth.2633.
Fontanelli G, Barate C, Ciabatti E, et al. Real-Time PCR and Droplet Digital PCR: two techniques for detection of the JAK2 mutation in Philadelphia-negative chronic myeloproliferative neoplasms. Int J Lab Hematol. 2015. doi:10.1111/ijlh.12404.
Idris S, Lindsay C, Kostiuk M, et al. Investigation of EZH2 pathways for novel epigenetic treatment strategies in oropharyngeal cancer. J Otolaryngol - Head Neck Surg. 2016;45(1):54. doi:10.1186/s40463-016-0168-9.
Watanabe M, Kawaguchi T, Isa SI, et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21(15):3552–60. doi:10.1158/1078-0432.CCR-14-2151.
Yu M, Carter KT, Makar KW, et al. MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles. Epigenetics. 2015;10(9):803–9. doi:10.1080/15592294.2015.1068490.
Wang P, Jing F, Li G, et al. Absolute quantification of lung cancer related microRNA by droplet digital PCR. Biosens Bioelectron. 2015;74:836–42. doi:10.1016/j.bios.2015.07.048.
Wang Q, Yang X, He Y, et al. Droplet digital PCR for absolute quantification of EML4-ALK gene rearrangement in lung adenocarcinoma. J Mol Diagn. 2015;17(5):515–20. doi:10.1016/j.jmoldx.2015.04.002.
Biron VL, Kostiuk M, Isaac A, et al. Detection of human papillomavirus type 16 in oropharyngeal squamous cell carcinoma using droplet digital polymerase chain reaction. Cancer. 2016. doi:10.1002/cncr.29976.
Kingma DW, Allen RA, Caughron SK, et al. Comparison of molecular methods for detection of HPV in oral and oropharyngeal squamous cell carcinoma. Diagn Mol Pathol. 2010;19(4):218–23. doi:10.1097/PDM.0b013e3181d0cd35.
Ahn SM, Chan JYK, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140(9):846–54. doi:10.1001/jamaoto.2014.1338.
Holland D, Hoppe-Seyler K, Schuller B, et al. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res. 2008;68(23):9964–72. doi:10.1158/0008-5472.CAN-08-1134.
Hyland PL, McDade SS, McCloskey R, et al. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J Virol. 2011;85(21):10999–1006. doi:10.1128/JVI.00160-11.
Lessa RC, Campos AHJFM, De Freitas CE, et al. Identification of upregulated genes in oral squamous cell carcinomas. Head Neck. 2013;35(10):1475–81. doi:10.1002/hed.23169.
Cao W, Feng Z, Cui Z, et al. Up-regulation of enhancer of zeste homolog 2 is associated positively with cyclin D1 overexpression and poor clinical outcome in head and neck squamous cell carcinoma. Cancer. 2012;118(11):2858–71. doi:10.1002/cncr.26575.
Perrone F, Gloghini A, Cortelazzi B, Bossi P, Licitra L, Pilotti S. Isolating p16-positive/HPV-negative oropharyngeal cancer: an effort worth making. Am J Surg Pathol. 2011;35(5):774–7. doi:10.1097/PAS.0b013e3182116a45. 8.
Schache AG, Liloglou T, Risk JM, et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res. 2011;17(19):6262–71. doi:10.1158/1078-0432.CCR-11-0388.
Robinson M, Schache A, Sloan P, Thavaraj S. HPV specific testing: a requirement for oropharyngeal squamous cell carcinoma patients. Head Neck Pathol. 2012;6 Suppl 1:83–90. doi:10.1007/s12105-012-0370-7.
Dalianis T, Grün N, Koch J, et al. Human papillomavirus DNA and p16(INK4a) expression in hypopharyngeal cancer and in relation to clinical outcome, in Stockholm, Sweden. Oral Oncol. 2015;51(9):857–61. doi:10.1016/j.oraloncology.2015.06.002.
Upile NS, Shaw RJ, Jones TM, et al. Squamous cell carcinoma of the head and neck outside the oropharynx is rarely human papillomavirus related. Laryngoscope. 2014;124(12):2739–44. doi:10.1002/lary.24828.
Friedrich RE, Sperber C, Jäkel T, Röser K, Löning T. Basaloid lesions of oral squamous epithelial cells and their association with HPV infection and P16 expression. Anticancer Res. 2010;30:1605–12.
Duray A, Descamps G, Decaestecker C, et al. Human papillomavirus DNA strongly correlates with a poorer prognosis in oral cavity carcinoma. Laryngoscope. 2012;122(7):1558–65. doi:10.1002/lary.23298.
Rettig EM, Wentz A, Posner MR, et al. Prognostic implication of persistent human papillomavirus type 16 DNA detection in oral rinses for human papillomavirus–related oropharyngeal carcinoma. JAMA Oncol. 2015;21205(7):907–15. doi:10.1001/jamaoncol.2015.2524.
Chuang AY, Chuang TC, Chang S, et al. Presence of HPV DNA in convalescent salivary rinses is an adverse prognostic marker in head and neck squamous cell carcinoma. Oral Oncol. 2008;44(10):915–9. doi:10.1016/j.oraloncology.2008.01.001.
Zhao M, Rosenbaum E, Carvalho AL, et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int J Cancer. 2005;117(4):605–10. doi:10.1002/ijc.21216.
Nordfors C, Vlastos A, Du J, et al. Human papillomavirus prevalence is high in oral samples of patients with tonsillar and base of tongue cancer. Oral Oncol. 2014;50(5):491–7. doi:10.1016/j.oraloncology.2014.02.012.
Fakhry C, Rosenthal BT, Clark DP, Gillison ML. Associations between oral HPV16 infection and cytopathology: evaluation of an oropharyngeal “pap-test equivalent” in high-risk populations. Cancer Prev Res (Phila). 2011;4(9):1378–84. doi:10.1158/1940-6207.CAPR-11-0284.
Funding
This research was supported by grants from the Alberta Head and Neck Center for Oncology and Reconstruction (AHNCOR) Foundation, the University of Alberta Hospital Foundation and the University of Alberta Department of Surgery Clinical Research Grant.
Authors’ contributions
AI helped design the study, recruited participants, collected patient specimens, analyzed the results, wrote the initial draft of the manuscript, and edited and approved the final version of the manuscript in its current form. MK performed the majority of ddPCR and RT-qPCR assays, oversaw many of the technical aspects of the study. HZ assisted in study design, recruited participants, collected patient specimens, and edited and approved the final version of the manuscript in its current form. CL assisted with RNA extractions and ddPCR. FM recruited participants, collected patient specimens, and edited and approved the final version of the manuscript in its current form. DAOC recruited participants, collected patient specimens, contributed to study design, and edited and approved the final version of the manuscript in its current form. JRH recruited participants, collected patient specimens, contributed to study design, and edited and approved the final version of the manuscript in its current form. DWJC recruited participants, collected patient specimens, contributed to study design, and edited and approved the final version of the manuscript in its current form. HS recruited participants, collected patient specimens, contributed to study design, and edited and approved the final version of the manuscript in its current form. VLB conceived the study, oversaw the design of the study, oversaw all clinical and technical aspects of the study, recruited participants, collected patient specimens, helped analyze the data, provided guidance in the initial draft of the manuscript, and edited and approved the final version of the manuscript in its current form. All authors read and approved the final manuscript.
Competing interests
None.
Consent for publication
Informed written consent was obtained from each participant involved in the study.
Ethics approval and consent to participate
Prior to commencement, ethics approval was obtained from the University of Alberta Health Research Ethics Board (Biomedical Panel) Pro00057994.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
Sensitivity of ddPCR and RT-qPCR for detection of target EEF2 and E7 RNA in serially diluted samples. (DOC 82 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Isaac, A., Kostiuk, M., Zhang, H. et al. Ultrasensitive detection of oncogenic human papillomavirus in oropharyngeal tissue swabs. J of Otolaryngol - Head & Neck Surg 46, 5 (2017). https://doi.org/10.1186/s40463-016-0177-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40463-016-0177-8