Abstract
Myelodysplasia (MDS) /myeloproliferative neoplasm (MPN) overlap syndrome has been described since the 2001 WHO classification as disorders that have both proliferative and dysplastic changes simultaneously. Specific disorders include chronic myelomonocytic leukemia (CMML), juvenile myelomonocytic leukemia (JMML), BCR-ABL negative atypical chronic myeloid leukemia (aCML) and unclassifiable MDS/MPN (MPN/MDS-U). Recurrent gene mutations in these conditions have been described. Among them, SETBP1 mutations have been identified in up to 32% of aCML, 24% of JMML, 18% of CMML and 10% of MDS/MPN-U patients. The mutation hotspot lies in the amino acid residues 858–871 in the SETBP1 protein. SETBP1 mutations in MDS/MPN overlap syndrome is associated with accelerated transformation to leukemia and poor prognosis. In this review, we summarized the latest data on the role of SETBP1 mutations in the overlap syndrome. SETBP1 mutations may serve as a biomarker for the diagnosis and poor prognosis of the overlap syndrome.
Similar content being viewed by others
Background
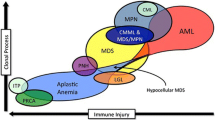
Myelodysplasia (MDS) /myeloproliferative neoplasm (MPN) overlap syndrome (MMOS) has been described as disorders that have both proliferative and dysplastic changes simultaneously [1,2,3,4,5]. Specific disorders include chronic myelomonocytic leukemia (CMML), juvenile myelomonocytic leukemia (JMML), BCR-ABL negative atypical chronic myeloid leukemia (aCML) and unclassifiable MDS/MPN (MPN/MDS-U). The overlap category of the disorders was first defined in the 2001 WHO classification of myeloid neoplasms [6] with the 2016 revision adding a fifth disorder – MDS/MPN with ringed sideroblasts and thrombocytosis (MDS/MPN-RS-T) [7]. Among MMOS, MDS/MPN-U represents about 2% of patients and usually carries a very poor prognosis with median survival of 21 months [8]. The definition of MDS/MPN-U has been updated in the 2008 version of WHO classifications [1]. A recent retrospective study validated the MDS-IPSS scoring system in MDS/MPN-U with low risk patients and delineated the natural history of this elusive malignancy [9].
There are frequent mutations in MDS, MPN, and acute myeloid leukemia (AML) [10,11,12,13,14,15,16,17]. Mutations in several pathways have been found to be useful biomarkers for diagnosis and some are druggable targets [18,19,20,21,22]. Ruxolitinib, enasidenib, midostaurin and several other small molecules are targeted agents in clinical applications and /or development for myeloid neoplasms [23,24,25,26,27,28]. However, no specific genetic mutations have been found for the MDS/MPN overlap syndrome. This review focused on the SETBP1 gene mutations and summarized the clinical correlations of the common mutations in MMOS and secondary AML (sAML).
SETBP1 mutations
SETBP1 was discovered in 2001 [29]. It is localized in chromosome 18q21.1. The cDNA encodes a protein with 1542 amino-acid residues. SETBP1 is a 170-kDa nuclear protein that binds to SET protein, a protein previously noted to be associated with acute undifferentiated leukemia [30].
Through whole exome sequencing of DNA from 727 patients with various myeloid malignancies, recurrent mutations of SETBP1 were found in 52 cases (7.2%) [31]. Among them, SETBP1 mutations have been identified in up to 32% of aCML, 24% of JMML, 18% of CMML and 10% of MDS/MPN-U patients. Somatic mutations of SETBP1 were associated with −7/del(7q) and poor prognosis, possibly due to its gain-of-function which promotes myeloid leukemic transformation in patients with myelodysplastic syndromes (MDS) and CMML. These data were consistent with those from other reports [32,33,34,35].
SETBP1 in AML transformation
The first clear link between SETBP1 and myeloid neoplasia was demonstrated in a patient with transformed AML [36]. The karyotyping studies performed at progression from MDS to AML in this patient revealed a t(12;18)(p13;q21) translocation leading to fusion between ETV6 & SETBP1. SETBP1 protects SET from protease cleavage. Overexpression of SETBP1 was shown to increase the level of SET protein, leading to increase in the formation of the SETBP1-SET-PP2A complex [36]. SETBP1 overexpression has been shown to confer growth advantage in hematopoietic progenitor cells, therefore promoting leukemogenic transformation. In a separate study of 386 patients with MDS, SETBP1 mutation was found to enhance ASXL1 mutation- induced differentiation block, and played a role as critical drivers in the leukemic transformation from MDS to AML [37].
SETBP1 mutations in MMOS
SETBP1 in CMML
One study using massive parallel sequencing investigated mutational hotspots of SETBP1 in 658 patients with MDS, CMML and sAML [38]. Among the 195 CMML patients included, 12 (6.2%) were positive for point mutations in SETBP1, a rate higher than those in MDS (2.2%) and sAML (1.7%). The mutation hotspot affected amino acid residues 858–871 in the SETBP1 protein. Those CMML patients with SETBP1 mutations had higher baseline WBC counts. SETBP1 mutations were more commonly associated with ASXL1 mutations and less commonly associated with TET2 mutations. After a median follow up of 31.1 months, SETBP1 mutated patients were noted to have worse OS [mOS: 21 months vs. 46.4 months; HR-2.23 (95% CI 1.07-4.26, p-0.028)]. Interestingly, when different CD34 positive cell fractions were genotyped by single-cell genome sequencing, all 4 potential driver mutations (ASXL1, SETBP1, ZRSR2 & SRSF2) occurred early in the hematopoietic stem cell populations. However, SETBP1 mutations were noted to occur after other driver mutations, and the mutant SETBP1 clone became predominant with progression to secondary AML [38]. The prevalence of SETBP1 mutations and their prognostic role in CMML were corroborated in other reports [39,40,41,42,43,44] (Table 1).
An independent prognostic scoring system for CMML, CPSS-Mol, was developed by incorporating clinical features and status of 38 commonly mutated genes in CMML [45]. The learning cohort consisted of 214 CMML patients, with 19 patients (8.9%) possessing SETBP1 mutations. Consistent with previous reports, SETBP1 mutations were associated with a higher WBC count (p < 0.001) and worse OS (HR for univariate: 2.49, p = 0.02; HR for multivariate: 2, p = 0.04). This scoring system was then validated in a cohort of 260 patients (24 patients possessed SETBP1 mutations; 9.2%). Based on the CPSS-Mol scores, the high-risk group had an overall survival of 17 months as opposed to mOS not yet reached in the low risk group (p < 0.001). This new scoring system may be useful in assisting clinical decision making and future clinical trials.
SETBP1 in juvenile myelomonocytic leukemia
SETBP1 mutation has also been reported in 4.8-7.2% of the patients with JMML [46, 47]. Most of the SETBP1 mutations co-existed with RAS pathway mutations with a lower allele burden, suggesting a secondary role in leukemogenesis [46]. Interestingly, this report also demonstrated an increasing allele burden with each round of intensive chemotherapy, suggestive of association of SETBP1 mutation with resistance to cytotoxic chemotherapy. Higher mutation rate was reported in additional studies [48, 49]. All these studies again confirmed poor prognosis in patients with SETBP1 mutations (Table 2).
SETBP1 in atypical CML
aCML is one of the heterogeneous neoplasms in the group of myelodysplastic/ myeloproliferative (MDS/MPN) overlap syndrome. Unlike CML, there is no diagnostic hallmark identified in aCML [50, 51]. Due to the poor understanding of the molecular pathogenesis of this disease, diagnosis has remained elusive and the prognosis is far worse than that of CML (median survival of 37 months after diagnosis) [52]. SETBP1 mutations was reported in 17/70 aCML patients (24%) using high throughput sequencing [34]. SETBP1 mutation was associated with worsened survival (22 months vs. 77 months, HR for death – 2.27, p = 0.01) and higher WBC counts (81 × 103 vs. 38.5 × 103 /□L, p = 0.008). In a separate study that included 60 aCML patients [32], 19/60 (32%) patients were noted to have SETBP1 mutations. SETBP1 appears to occur with significant frequency in aCML with negative impact on prognosis. SETBP1 mutations may be a new biomarker for aCML diagnosis and prognosis (Table 3).
SETBP1 in MDS/MPN-U
In a large study of 1130 patients with myeloid neoplasms, SETBP1 mutations were found in 20/240 (9.3%) MDS/MPN-U patients [32]. SETBP1 mutated (mSETBP1) patients had significantly higher WBC counts and lower platelet counts and hemoglobin levels than those patients with wide-type SETBP1. Dysplastic morphology was more common in mSETBP1 cases as compared with wild-type cases. This study also reported a significant association of SETBP1 mutations with isochromosome i(17)(q10). The similar findings were reported by a separate group [53]. These two studies also concurred that SETBP1 mutations were strongly associated with ASXL1 mutation. Therefore, SETBP1 mutations can serve as a new biomarker for MDS/MPN-U (Table 3).
Conclusion
SETBP1 mutations are relatively common in MDS/MPN overlap syndrome. The mutation hotspot lies in the amino acid residues 858–871 in the SETBP1 protein. SETBP1 overexpression is associated with accelerated transformation to leukemia and poor prognosis. SETBP1 mutations may serve as a biomarker for the diagnosis and poor prognosis for the overlap syndrome.
Abbreviations
- aCML:
-
atypical chronic myeloid leukemia
- CMML:
-
Chronic myelomonocytic leukemia
- JMML:
-
Juvenile myelomonocytic leukemia
- MDS:
-
Myelodysplasia
- MPN:
-
Myeloproliferative neoplasm
- MPN/MDS-U:
-
Unclassifiable MDS/MPN
References
Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51.
Geyer HL, Mesa RA. Therapy for myeloproliferative neoplasms: when, which agent, and how? Blood. 2014;124(24):3529–37.
Kaifie A, Kirschner M, Wolf D, Maintz C, Hänel M, Gattermann N, Gökkurt E, Platzbecker U, Hollburg W, Göthert JR, Parmentier S, Lang F, Hansen R, Isfort S, Schmitt K, Jost E, Serve H, Ehninger G, Berdel WE, Brümmendorf TH, Koschmieder S. Bleeding, thrombosis, and anticoagulation in myeloproliferative neoplasms (MPN): analysis from the German SAL-MPN-registry. J Hematol Oncol. 2016;9(1):18.
Mehta J, Wang H, Iqbal SU, Mesa R. Epidemiology of myeloproliferative neoplasms in the United States. Leuk Lymphoma. 2014;55(3):595–600.
Feng G, Gale RP, Cui W, Cai W, Huang G, Xu Z, Qin T, Zhang Y, Li B, Fang L, Zhang H, Pan L, Hu N, Qu S, Wang J, Cui Y, Xiao Z. A systematic classification of megakaryocytic dysplasia and its impact on prognosis for patients with myelodysplastic syndromes. Exp Hematol Oncol. 2016;5(1):12.
Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–302.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405.
Orazi A, Germing U. The myelodysplastic/myeloproliferative neoplasms: myeloproliferative diseases with dysplastic features. Leukemia. 2008;22(7):1308–19.
DiNardo CD, Daver N, Jain N, Pemmaraju N, Bueso-Ramos C, Yin CC, Pierce S, Jabbour E, Cortes JE, Kantarjian HM, Garcia-Manero G, Verstovsek S. Myelodysplastic/myeloproliferative neoplasms, unclassifiable (MDS/MPN, U): natural history and clinical outcome by treatment strategy. Leukemia. 2014;28(4):958–61.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–9.
Tirado CA, Siangchin K, Shabsovich DS, Sharifian M, Schiller G. A novel three-way rearrangement involving ETV6 (12p13) and ABL1 (9q34) with an unknown partner on 3p25 resulting in a possible ETV6-ABL1 fusion in a patient with acute myeloid leukemia: a case report and a review of the literature. Biomark Res. 2016;4(1):16.
Hwang SM, Kim SY, Kim JA, Park H-S, Park SN, Im K, Kim K, Kim S-M, Lee DS. Short telomere length and its correlation with gene mutations in myelodysplastic syndrome. J Hematol Oncol. 2016;9(1):62.
Gao J, Chen Y-H, Peterson LC. GATA family transcriptional factors: emerging suspects in hematologic disorders. Exp Hematol Oncol. 2015;4(1):28.
Laszlo GS, Alonzo TA, Gudgeon CJ, Harrington KH, Kentsis A, Gerbing RB, Wang Y-C, Ries RE, Raimondi SC, Hirsch BA, Gamis AS, Meshinchi S, Walter RB. High expression of myocyte enhancer factor 2C (MEF2C) is associated with adverse-risk features and poor outcome in pediatric acute myeloid leukemia: a report from the Children’s oncology group. J Hematol Oncol. 2015;8(1):115.
Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373(12):1136–52.
Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD, Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP, Teague JW, O'Meara S, McLaren S, Bianchi M, et al. Somatic CALR mutations in Myeloproliferative Neoplasms with nonmutated JAK2. N Engl J Med. 2013;369(25):2391–405.
Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, Shlien A, Groves MJ, Forbes SA, Raine K, Hinton J, Mudie LJ, McLaren S, Hardy C, Latimer C, Della Porta MG, O'Meara S, Ambaglio I, Galli A, Butler AP, Walldin G, Teague JW, Quek L, Sternberg A, Gambacorti-Passerini C, Cross NC, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–27.
Tibes R, Al-Kali A, Oliver GR, Delman DH, Hansen N, Bhagavatula K, Mohan J, Rakhshan F, Wood T, Foran JM, Mesa RA, Bogenberger JM. The hedgehog pathway as targetable vulnerability with 5-azacytidine in myelodysplastic syndrome and acute myeloid leukemia. J Hematol Oncol. 2015;8(1):114.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, Gundem G, Van Loo P, Martincorena I, Ganly P, Mudie L, McLaren S, O’Meara S, Raine K, Jones DR, Teague JW, Butler AP, Greaves MF, Ganser A, Döhner K, Schlenk RF, Döhner H, Campbell PJ. Genomic classification and prognosis in acute myeloid Leukemia. N Engl J Med. 2016;374(23):2209–21.
Geyer HL, Mesa RA. Therapy for myeloproliferative neoplasms: when, which agent, and how? Hematology Am Soc Hematol Educ Program. 2014;2014(1):277–86.
Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, Wilson RK, Baty JD, Duncavage EJ, Tandon B, Lee Y-S, Wartman LD, Uy GL, Ghobadi A, Tomasson MH, Pusic I, Romee R, Fehniger TA, Stockerl-Goldstein KE, Vij R, Oh ST, Abboud CN, Cashen AF, Schroeder MA, Jacoby MA, Heath SE, Luber K, Janke MR, Hantel A, Khan N, et al. TP53 and Decitabine in acute myeloid Leukemia and Myelodysplastic syndromes. N Engl J Med. 2016;375(21):2023–36.
Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, Habdank M, Späth D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A, Döhner H. Mutations and treatment outcome in cytogenetically normal acute myeloid Leukemia. N Engl J Med. 2008;358(18):1909–18.
Yacoub A, Prochaska L. Ruxolitinib improves symptoms and quality of life in a patient with systemic mastocytosis. Biomark Res. 2016;4(1):2.
Maffini E, Giaccone L, Festuccia M, Brunello L, Buondonno I, Ferrero D, Boccadoro M, Dellacasa C, Busca A, Novero D, Bruno B. Ruxolitinib in steroid refractory graft-vs.-host disease: a case report. J Hematol Oncol. 2016;9(1):67.
Verstovsek S, Odenike O, Singer JW, Granston T, Al-Fayoumi S, Deeg HJ. Phase 1/2 study of pacritinib, a next generation JAK2/FLT3 inhibitor, in myelofibrosis or other myeloid malignancies. J Hematol Oncol. 2016;9(1):137.
Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Dohner K, Marcucci G, Lo-Coco F, Klisovic RB, Wei A, Sierra J, Sanz MA, Brandwein JM, de Witte T, Niederwieser D, Appelbaum FR, Medeiros BC, Tallman MS, Krauter J, Schlenk RF, Ganser A, Serve H, Ehninger G, Amadori S, Larson RA, Dohner H. Midostaurin plus chemotherapy for acute myeloid Leukemia with a FLT3 mutation. N Engl J Med. 2017;377(5):454–64.
Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW, Kantarjian HM, Collins R, Patel MR, Frankel AE, Stein A, Sekeres MA, Swords RT, Medeiros BC, Willekens C, Vyas P, Tosolini A, Xu Q, Knight RD, Yen KE, Agresta S, de Botton S, Tallman MS. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722–31.
Amatangelo MD, Quek L, Shih A, Stein EM, Roshal M, David MD, Marteyn B, Farnoud NR, de Botton S, Bernard OA, Wu B, Yen KE, Tallman MS, Papaemmanuil E, Penard-Lacronique V, Thakurta A, Vyas P, Levine RL. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 2017;130(6):732–41.
Minakuchi M, Kakazu N, Gorrin-Rivas MJ, Abe T, Copeland TD, Ueda K, Adachi Y. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268(5):1340–51.
Adachi Y, Pavlakis GN, Copeland TD. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J Biol Chem. 1994;269(3):2258–62.
Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, Ng KP, Gudmundsson KO, Vishwakarma BA, Jerez A, Gomez-Segui I, Takahashi M, Shiraishi Y, Nagata Y, Guinta K, Mori H, Sekeres MA, Chiba K, Tanaka H, Muramatsu H, Sakaguchi H, Paquette RL, McDevitt MA, Kojima S, Saunthararajah Y, Miyano S, Shih LY, Du Y, Ogawa S, Maciejewski JP. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45(8):942–6.
Meggendorfer M, Bacher U, Alpermann T, Haferlach C, Kern W, Gambacorti-Passerini C, Haferlach T, Schnittger S. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. 2013;27(9):1852–60.
Patnaik MM, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, Knudson RA, Ketterling RP, Tefferi A, Solary E. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28(11):2206–12.
Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, Antolini L, Mologni L, Donadoni C, Papaemmanuil E, Schnittger S, Kim DW, Boultwood J, Rossi F, Gaipa G, De Martini GP, di Celle PF, Jang HG, Fantin V, Bignell GR, Magistroni V, Haferlach T, Pogliani EM, Campbell PJ, Chase AJ, Tapper WJ, Cross NC, Gambacorti-Passerini C. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45(1):18–24.
Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, Wlodarski MW, Kolking B, Wichmann M, Gorlich K, Gohring G, Bug G, Ottmann O, Niemeyer CM, Hofmann WK, Schlegelberger B, Ganser A, Heuser M. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–84.
Cristobal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, Novo FJ, Bandres E, Calasanz MJ, Bernabeu C, Odero MD. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115(3):615–25.
Inoue D, Kitaura J, Matsui H, Hou HA, Chou WC, Nagamachi A, Kawabata KC, Togami K, Nagase R, Horikawa S, Saika M, Micol JB, Hayashi Y, Harada Y, Harada H, Inaba T, Tien HF, Abdel-Wahab O, Kitamura T. SETBP1 mutations drive leukemic transformation in ASXL1-mutated MDS. Leukemia. 2015;29(4):847–57.
Damm F, Itzykson R, Kosmider O, Droin N, Renneville A, Chesnais V, Gelsi-Boyer V, de Botton S, Vey N, Preudhomme C, Clavert A, Delabesse E, Park S, Birnbaum D, Fontenay M, Bernard OA, Solary E. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013;27(6):1401–3.
Laborde RR, Patnaik MM, Lasho TL, Finke CM, Hanson CA, Knudson RA, Ketterling RP, Pardanani A, Tefferi A. SETBP1 mutations in 415 patients with primary myelofibrosis or chronic myelomonocytic leukemia: independent prognostic impact in CMML. Leukemia. 2013;27(10):2100–2.
Cui Y, Tong H, Du X, Li B, Gale RP, Qin T, Liu J, Xu Z, Zhang Y, Huang G, Jin J, Fang L, Zhang H, Pan L, Hu N, Qu S, Xiao Z. Impact of TET2, SRSF2, ASXL1 and SETBP1 mutations on survival of patients with chronic myelomonocytic leukemia. Exp Hematol Oncol. 2015;4:14.
Patnaik MM, Parikh SA, Hanson CA, Tefferi A. Chronic myelomonocytic leukaemia: a concise clinical and pathophysiological review. Br J Haematol. 2014;165(3):273–86.
Patnaik MM, Wassie EA, Padron E, Onida F, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, Ketterling RP, Komrokji R, Tefferi A, Solary E. Chronic myelomonocytic leukemia in younger patients: molecular and cytogenetic predictors of survival and treatment outcome. Blood Cancer J. 2015;5:e270.
Shou LH, Cao D, Dong XH, Fang Q, Wu Y, Zhang Y, Fei JP, Xu BL. Prognostic significance of SETBP1 mutations in myelodysplastic syndromes, chronic myelomonocytic leukemia, and chronic neutrophilic leukemia: a meta-analysis. PLoS One. 2017;12(2):e0171608.
Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, Kohlmann A, Alpermann T, Yoshida K, Ogawa S, Koeffler HP, Kern W, Haferlach C, Schnittger S. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood. 2012;120(15):3080–8.
Elena C, Galli A, Such E, Meggendorfer M, Germing U, Rizzo E, Cervera J, Molteni E, Fasan A, Schuler E, Ambaglio I, Lopez-Pavia M, Zibellini S, Kuendgen A, Travaglino E, Sancho-Tello R, Catricala S, Vicente AI, Haferlach T, Haferlach C, Sanz GF, Malcovati L, Cazzola M. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128(10):1408–17.
Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, Kon A, Sanada M, Chiba K, Tanaka H, Makishima H, Wang X, Xu Y, Doisaki S, Hama A, Nakanishi K, Takahashi Y, Yoshida N, Maciejewski JP, Miyano S, Ogawa S, Kojima S. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013;45(8):937–41.
Shiba N, Ohki K, Park MJ, Sotomatsu M, Kudo K, Ito E, Sako M, Arakawa H, Hayashi Y. SETBP1 mutations in juvenile myelomonocytic leukaemia and myelodysplastic syndrome but not in paediatric acute myeloid leukaemia. Br J Haematol. 2014;164(1):156–9.
Stieglitz E, Troup CB, Gelston LC, Haliburton J, Chow ED, Yu KB, Akutagawa J, Taylor-Weiner AN, Liu YL, Wang YD, Beckman K, Emanuel PD, Braun BS, Abate A, Gerbing RB, Alonzo TA, Loh ML. Subclonal mutations in SETBP1 confer a poor prognosis in juvenile myelomonocytic leukemia. Blood. 2015;125(3):516–24.
Bresolin S, De Filippi P, Vendemini F, D'Alia M, Zecca M, Meyer LH, Danesino C, Locatelli F, Masetti R, Basso G, Te Kronnie G. Mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia: a report from the Italian AIEOP study group. Oncotarget. 2016;7(20):28914–9.
Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, Bottomly D, Wilmot B, McWeeney SK, Tognon CE, Pond JB, Collins RH, Goueli B, Oh ST, Deininger MW, Chang BH, Loriaux MM, Druker BJ, Tyner JW. Oncogenic CSF3R mutations in chronic Neutrophilic Leukemia and atypical CML. N Engl J Med. 2013;368(19):1781–90.
Wang SA, Hasserjian RP, Fox PS, Rogers HJ, Geyer JT, Chabot-Richards D, Weinzierl E, Hatem J, Jaso J, Kanagal-Shamanna R, Stingo FC, Patel KP, Mehrotra M, Bueso-Ramos C, Young KH, Dinardo CD, Verstovsek S, Tiu RV, Bagg A, Hsi ED, Arber DA, Foucar K, Luthra R, Orazi A. Atypical chronic myeloid leukemia is clinically distinct from unclassifiable myelodysplastic/myeloproliferative neoplasms. Blood. 2014;123(17):2645–51.
Gotlib J. How I treat atypical chronic myeloid leukemia. Blood. 2017;129(7):838–45.
Kanagal-Shamanna R, Luthra R, Yin CC, Patel KP, Takahashi K, Lu X, Lee J, Zhao C, Stingo F, Zuo Z, Routbort MJ, Singh RR, Fox P, Ravandi F, Garcia-Manero G, Medeiros LJ, Bueso-Ramos CE. Myeloid neoplasms with isolated isochromosome 17q demonstrate a high frequency of mutations in SETBP1, SRSF2, ASXL1 and NRAS. Oncotarget. 2016;7(12):14251–8.
Acknowledgements
None.
Funding
There was no funding involved in this study.
Availability of data and materials
All data are published in the study.
Author information
Authors and Affiliations
Contributions
DL designed the study. All authors drafted and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This is not applicable.
Consent for publication
This is not applicable.
Competing interests
The authors have no relevant competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Linder, K., Iragavarapu, C. & Liu, D. SETBP1 mutations as a biomarker for myelodysplasia /myeloproliferative neoplasm overlap syndrome. Biomark Res 5, 33 (2017). https://doi.org/10.1186/s40364-017-0113-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40364-017-0113-8