Abstract
Background
There has been a growing interest in camel anaplasmosis due to its recent emergence in this reservoir species and concerns for its zoonotic potential. The epidemiology of anaplasmosis in camels therefore remains poorly understood mostly because camels belong to marginalised poor and often transhumant populations whose interests are largely neglected. Most studies of anaplasmosis in camels have relied on microscopy and serology for diagnosis and only three studies, undertaken in Tunisia, Saudia Arabia and China, have used molecular diagnostics. The present work characterises Anaplasmataceae strains circulating in the Camelus dromedarius reservoir in Morocco using PCR.
Methods
Camels (n = 106) were randomly sampled from 6 regions representing different agro-ecological areas in southern Morocco. Whole blood was collected and screened using PCR methods targeting the gene groEL. Anaplasmataceae strains were characterised by sequence analysis of the gene groEL.
Results
A total of 39.62% (42/106) camels screened were positive for Anaplasmataceae spp. GenBank BLAST analysis of five positive sequenced samples revealed that all strains were 100% identical to “Candidatus Anaplasma camelii”. Phylogenetic investigation and genetic characterisation of the aligned segment (650 bp) of the gene groEL confirmed high similarity with A. platys.
Conclusion
This study demonstrates the circulation of a previously unidentified species of the genus Anaplasma in Morocco which is genetically close to the agent causing canine anaplasmosis but whose main reservoir is thought to be Camelus dromedarius.
Trial registration number
This study is not a clinical trial and therefore a trial registration number does not apply.
Similar content being viewed by others
Multilingual abstracts
Please see Additional file 1 translations of the abstract into the six official working languages of the United Nations.
Background
Tick-borne diseases, especially those caused by Rickettsiae, are a major source of economic burden for livestock keepers due to their impact on productivity in ruminant hosts and that several pathogens in this group are also zoonotic. Anaplasmosis is tick borne and caused by gram negative, obligate intracellular bacteria of the genus Anaplasma [1]. The epidemiology of anaplasmosis is complex due to the diversity of Anaplasma species that cause the condition, the wide host range and the role of a vector in its transmission.
The Anaplasma genus includes, but is not limited to, the following species: (1) A. marginale, (2) A. centrale, (3) A. ovis, (4) A. bovis, (5) A. platys, and (6) A. phagocytophilum. (1) A. marginale is the aetiological agent of bovine intra-erythrocytic anaplasmosis [2]. Infection occurs through the bite of a tick carrying the bacteria [3, 4]. Hard ticks, including Rhipicephalus spp., Boophilus spp., Dermacentor spp. and Ixodes ricinus are the main source of transmission, although other sources of biological and mechanical transmission have been reported [5, 6]. Post infection, the incubation lasts for 7 to 60 days after which if parasitaemia of red blood cells exceeds the 15% threshold, clinical signs appear [7, 8]. The severity of signs observed during the clinical phase varies depending on strain virulence and immune status of infected cattle. In general, infected cattle present with anaemia, pyrexia, lethargy, weight loss, milk drop in lactating females and occasionally abortion for in-calf cows. Death may occur in the absence of chemotherapy and veterinary care [7, 9]. (2) A. centrale preferentially infects cattle and is used as a live vaccine against A. marginale in cattle in Australia, South Africa and South America because of its lower virulence and good cross immunity [10]. Small ruminants are preferentially infected by A. ovis (3) and prevalence has been reported to be high in several countries [11–13] with considerable economic impact [13]. Clinical cases usually present in stressed, immune-depressed sheep and goats or in cases of co-infection with clinical signs similar to those observed for A. marginale infected cattle [14, 15]. A. ovis transmission to small ruminants occurs through tick bites as described for cattle, although Rhipicephalus spp. play a greater role [14].
In addition to intra-erythrocytic Anaplasma species, the genus also includes A. bovis (4), which causes intra-monocytic anaplasmosis, a sub-clinical or benign clinical form of the disease [16]. Other species include A. platys (5), which has a tropism for platelets in dogs and causes canine cyclic thrombocytopenia [17] and A. phagocytophilum (6) which causes tick-borne fever (TBF) in domestic ruminants [18], granulocytic anaplasmosis (GA) in humans [19], Equine GA in horses [20], canine GA in dogs [21] and feline GA in cats [22]. Like A. phagocytophilum, A. ovis has been found on rare occasions to be zoonotic [23, 24].
Despite the limited number of studies undertaken on anaplasmosis in camels, evidence to date would suggest that one-humped camels (Camelus dromedarius) are not a preferential host for the Anaplasma. The only Anaplasma species found in this camel are genetically related to A. platys [25–27]. BenSaid et al. (2014) [28] reported A. phagocytophilum seropostive camels in Tunisia but this serological diagnosis was not confirmed by molecular methods.
During the last three years, an outbreak of undiagnosed disease in camels causing clinical signs of dependant oedema, anorexia, respiratory distress and sudden death was reported in the southern regions of Morocco by livestock keepers and veterinary services. The presentation of this undiagnosed illness was similar to the clinical signs observed in cattle acutely infected with A. phagocytophilum and given the practice of trans-boundary transhumance across the Sahara of North Africa it was thought likely that camel anaplasmosis would be present in Morocco. The present study investigates and characterises Anaplasmataceae spp. infection in Camelus dromedarius in Morocco using molecular tools.
Methods
Region and study population
A cross-sectional survey was undertaken between December 2013 and April 2015 with camel herds were purposefully selected based on owner willingness to participate in the study. Sampling was conducted across 37 sites in six regions of southern Morocco including areas where the outbreak of undiagnosed disease was reported (Fig. 1). 106 camels were sampled in total. At the herd level, a sub-sample of camels was randomly tested. Four of the camels sampled showed signs of dependant oedema at the time of sampling (Fig. 2). Whole blood was collected from the jugular vein using EDTA vacutainers® and was subsequently aliquoted and stored at −20 °C until further analysis. Ticks were collected from camels and were identified using standard keys [29].
DNA extraction
DNA was extracted from camel whole blood using the kit NucleoSpin® Blood Quick Pure (Macherey-Nagel, Düren, Germany) according to manufacturer instructions. DNA was stored at −20 °C until amplification.
Polymerase chain reaction
All extracted DNA was amplified targeting the ‘heat-shock operon’ ‘groEL’ using Anaplasmataceae-specific PCR primers AnaplatF2 5’-GCGTAGTCCGATTCTCCAGT-3’ and AnaGro712R 5’-CCGCGATCAAACTGCATACC-3’ [25, 30]. A final PCR mix volume of 25 μl was prepared by adding 1.5 μl of each primer, 12.5 μl of Taq DNA Promega GoTaq® Hot Start Colorless Master Mix (Promega corporation, Madison,WI, USA), 4.5 μl DNA-free water and 5 μl of DNA to amplify. The thermocycler Eppendorf Mastercycler® (Eppendorf, Hamburg, Germany) was programmed for an initial denaturation at 95 °C during 8 min, followed by 35 cycles of denaturation at 94 °C during 1 min, hybridisation at 59 °C during 40 s and elongation at 72 °C during 1 min. The programme ended with a final extension at 72 °C during 10 min.
Samples found to be positive by Anaplasmataceae specific PCR were screened for A. phagocytophilum using forward and reverse primers -903f 5’-AGTTTGACTGGAACACACCTGATC-3’ and 1024r 5’-CTCGTAACCAATCTCAAGCTCAAC-3’ targeting a portion of the msp2 gene (122 bp) [31]. The master mix was prepared as described above. Amplification started with an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 20 s, hybridisation at 50 °C for 30 s, elongation at 72 °C for 1 min and a final elongation at 72 °C for 10 min. A reference positive and negative sample were incorporated and amplified for each PCR.
PCR products were subsequently visualised through electrophoresis in 2% agarose gel using SYBR® Safe DNA gel stain (Invitrogen, Carlsbad, USA). The band size of the amplicon of interest was 650 bp.
Purification, sequencing and phylogenetic analysis
Five positive PCR products of Anaplasmataceae spp. were selected for purification using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). The purified DNA was then sequenced in both directions using the same primers as those used for PCR. Sequencing was performed by Inserm (Institut Cochin, 22, rue Mechain, 75 014 Paris, France; http://cochin.inserm.fr/les-plateformes/genomique-et-transcriptomique/plate-forme-de-sequencage/activite-sequencage).
GenBank BLAST analysis was used to compare the sequences obtained to those of reference strains. The algorithm ClustalW® and BioEdit® software were used to align multiple sequences. The dendrogram was constructed using the Mega software and the Neighbour joining (NJ) method for distance, parwise deletion and the boostrap test with 1,000 reiterations.
The software WinPepi® v11.42 [32] was used to compute the chi-square statistic.
Results
39.62% (42/106) of camels showed a PCR band corresponding to the size of the nucleotide portion of the groEL target gene. The Oued Ed-Dahab region was found to have the highest percentage of positives, with 50.00% (9/18) of camels affected and the lowest rate in the region of Es-Semara, with only 12.50% (2/16). Using the Pearson chi-square test, the difference between the inter-regional prevalence was not found to be statistically significant (Chi-square 6 193, P > 0.05).
All four camels showing dependant oedema were found to be positive. Sampled camels were found to be exclusively infested with the tick species Hyalomma dromedarii. None of the 42 samples analysed using PCR targeting msp2 gene was found to be positive for A. phagocytophilum. The results are summarised in Table 1.
Out of the 42 Anaplasmataceae spp. PCR positive samples, one sample was randomly selected from each region giving a total of five samples sequenced with only one variant of Anaplasma sp. was found. The partial groEL sequences of Anaplasma sp. strains of the same lineage to “Candidatus Anaplasma camelii” isolated from camels were all 100% identical. One of these sequences was submitted to GenBank (GenBank accession number: KX074079).
GenBank BLAST analysis confirmed that these sequences were 100% genetically identical to “Candidatus Anaplasma camelii” [GenBank accession number: KJ814955] camel strains from Saudi Arabia [25]. Similarity with A. platys varied between 89% and 93% [GenBank accession number: AY008300, EU004824, EU004825, HQ718723, JN121382 and EU516386] and similarity with A. phagocytophilum between 83 and 84% [GenBank accession number: AY279085, JX133175].
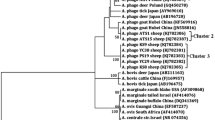
Phylogenetic analysis of the 650 bp portion of groEL was undertaken for GenBank reference strains and study strains. Study sequences were found to cluster with those of “Candidatus Anaplasma camelii” [GenBank accession number: KJ814955, KJ814957-KJ814959] and to have a closer genetic lineage to A. platys than any other Anaplasma species. The high weight index of nodes obtained using the bootstrap-test as well as the identical amino acid sequences confirm this result (Fig. 3).
Phylogenetic tree of the groEL gene derived from Moroccan camel strains of Anaplasma sp. The tree was obtained using the neighbour joining method with software Mega after alignment with ClustalW of 650 bp sequences of the groEL gene from this study and Anaplasma sp. sequences available from GenBank from various host species and countries of origin. We used Kimura 2-parameter method to calculate distance matrices. In each node, percentages of bootstrap values (1 000 repeats) are indicated
Discussion
Camelidae are described as ‘pseudo’ ruminants and are distributed mostly in semi-arid and arid zones of Africa, Middle East and Asia. They are recognised for their ability to resist and thrive in the extreme and unfavourable conditions typical of these arid areas. Camel keeping is an important livelihood for populations of the southeast and southwest regions of Morocco providing income and provision of milk and meat. They are also used to transport nomads and their belongings during transhumance as well as traction.
There is a dearth of evidence on tick-borne (and other) diseases in camels, primarily because camels are kept by poor marginalised nomadic populations. Camel health is of limited interest to pharmaceutical companies because of perceived low profit-margins of drug sales and veterinary interventions. On a global scale, there are only a few studies on this group of pathogens in this species. This is in part due to the difficulty in accessing nomadic camel-keeping populations, by virtue of the fact that camels are few in number and dispersed across the immense area of the Sahara Desert. This is the first study on anaplasmosis in camels in the Kingdom of Morocco.
Anaplasmataceae spp. was found in 35.85% of ‘apparently healthy’ camels not showing clinical signs at the time of sampling; four (9.5%) of the 42 PCR-positive animals had clinical signs of infection. Camels sampled in this study were shown to be infected by a single variant of Anaplasma sp. identical to “Candidatus Anaplasma camelii” identified and named by Bastos et al. [25] for the same host species in Saudi Arabia. The related strain A. platys is known to cause cyclic thrombocytopenia in dogs. However, the pathogenicity of “Candidatus Anaplasma camelii” in camels and other species and its zoonotic potential are unknown. Nevertheless, it is important to note that sampling coincided with an outbreak of undiagnosed disease in 2013 onwards in Moroccan camels.
The four camels showing clinical signs of the previously undiagnosed disease came from different regions (Provinces of Laâyoune, Es-Semara, Boujdour and Oued Ed Dahab). The initial clinical sign observed was limited to dependent oedema that spread to the whole body, at which point the animals became recumbent and eventually died. In the absence of more in-depth diagnostic investigations, and given that trypanosomiasis is probably endemic in these regions [33, 34], the cause of this syndrome remains uncertain. As all four camels showing clinical signs were found to be PCR positive, we can suggest but not confirm that “Candidatus Anaplasma camelii” infection would have contributed in the clinical signs observed. Out of the five positive samples randomly selected and sequenced, only one was from the group of four camels showing clinical signs i.e. four of the sequenced samples were from apparently healthy camels; of the other three camels showing clinical signs all were positive for Anaplamataceae spp. but identification to species level will require further sequencing. Intra-erythrocytic anaplasmosis due to A. marginale is the only Anaplasma species confirmed to cause sub-clinical disease in camels [35].
Several studies have used the groEL gene ton discriminate between Anaplasma species [25, 30, 36, 37]. Sequencing of the groEL gene differentiated Anaplasma sp. variants circulating in camels and A. platys in dogs - which formed a cluster - and all other forms of anaplasmosis in different animal species - which formed a separate cluster (Fig. 3). Confirmation of infection with “Candidatus Anaplasma camelii” is limited to the five strains sequenced because (i) the groEL primers used to screen the samples, whilst preferentially amplifying Anaplasma species, are not specific to Anaplasma and (ii) without sequencing all amplicons one cannot conclude that all PCR positive samples contain “Candidatus Anaplasma camelii”. In a previous study which made use of the same primers [25], the presence of an Ehrlichia strain (closely related to E. canis) was detected in 3% of the dromedary camels from Saudi Arabia. As just five amplicons were selected for sequencing in this study of camels from Morocco, it is possible that Ehrlichia strains may also be present in camels from Morocco, and/or that multiple Anaplasma species may be present.
This study has shown that camels in Morocco are probably a reservoir for “Candidatus Anaplasma camelii” vectored by the tick Hyalomma spp. that becomes infected during its larval and nymphal stage by feeding on small desert animals. After maturation to the adult stage, the tick then inoculates the bacteria when taking a blood meal from larger mammals (e.g. camels, dogs and wild desert mammals such as the jackal).
Transmission to a range of domestic hosts is likely due to livestock management practices. As part of the transhumant lifestyle, camels, small ruminants and dogs belonging to the same owner are managed as a single unit. During the dry season, watering holes (known as Guelta) become severely limited and livestock keepers congregate with their livestock increasing risk of transmission as camel breeders in these areas rarely use preventive measures against ticks.
Prevalence values reported for other countries are lower than the 39.62% obtained here. A study by Bastos et al. [25] undertaken in Saudi Arabia using PCR targetting 16 s rRNA genes and the groEL gene reported a camel anaplasmosis prevalence of 26%. In this study no camels were found to be positive for A. phagocytophilum. However in Tunisia, a neighbouring country with similar camels rearing conditions and practices to those of Morocco, 29.2% of camels were seropositive for A. phagocytophilum [28]. This has to be interpreted with caution as cross-reactions with other Anaplasma species can occur [38, 39]. A later study re-screened blood samples from the same animals by PCR (16S rRNA) and prevalence was 17.7%. The strain in these Tunisian camels was characterised as Anaplasma sp. and considered as A. platys-like [26]. In the Canary Islands, anti Anaplasma sp. antibodies were detected in 3% of camels sampled [40]. In China, A. platys infections have been reported in clinically healthy Bactrian camels with 7.2% prevalence [27]. In Nigeria, prevalence ranged from 3.8 to 16.5% for intra-erythrocytic anaplasmosis due to A. marginale in camels [41–44]. However, infection of camels with A. marginale has never been confirmed using molecular methods, either in Nigeria or elsewhere in the world.
Moreover, In agreement to what has been reported in Tunisia [26, 28], camels sampled in this study were mainly infested with hard ticks of Hyalomma sp. genus and not Ixodes ricinus. Heavy tick infestations were mostly observed in juvenile animals, which are infested with nymphs, referred to colloquially as Delma. This triggers intense pruritus and hair loss (Figs. 4 and 5), anaemia through mass tick feeding and eventually death if infected with a tick-borne disease.
In the Maghreb region, the tick genus Hyalomma sp. has been reported to be a potential vector of several pathogens, including Rickettsia aeschlimannii, R. africae [45–47], Ehrlichia sp. and A. phagocytophilum [48]. This tick is widely distributed in southern Morocco [49] as it is adapted to the extreme desert conditions, and it has been demonstrated to infest a large range of animals [50, 51], suggesting that Hyalomma sp. could also be a vector for “Candidatus Anaplasma camelii”.
Conclusions
This is the first report of camel anaplasmosis in Morocco and to genetically characterise “Candidatus Anaplasma camelii”. Infection of camels with this bacterium does not seem to cause clinical disease, but the high prevalence would suggest that camels are the principal host of this pathogen. Further studies are required to determine: (i) the role of different vectors (tick and insect) in transmission (ii) the role of domestic and wild species as reservoir or dead-end hosts (iii) the zoonotic potential of this pathogen and (iv) its pathogenicity in camels.
Abbreviations
- A:
-
Anaplasma
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- GA:
-
Granulocytic anaplasmosis
- HGA:
-
Human granulocytic anaplasmosis
- PCR:
-
Polymerase chain reaction
- rRNA:
-
Ribosomal ribonucleic acid
- TBF:
-
Tick-borne fever
References
Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families rickettsiaceae and anaplasmataceae in the order rickettsiales: unification of some species of ehrlichia with anaplasma, cowdria with ehrlichia and ehrlichia with neorickettsia, descriptions of six new species combinations and designation of ehrlichia equi and “HGE agent” as subjective synonyms of ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–65.
Aubry P, Geale DW. A review of bovine anaplasmosis. Transbound Emerg Dis. 2011;58:1–30.
Kocan KM, Goff WL, Stiller D, Claypool PL, Edwards W, Ewing SA, et al. Persistence of anaplasma marginale (rickettsiales: anaplasmataceae) in male Dermacentor andersoni (acari: ixodidae) transferred successively from infected to susceptible calves. J Med Entomol. 1992;29:657–68.
Kocan KM, Stiller D, Goff WL, Claypool PL, Edwards W, Ewing SA, et al. Development of anaplasma marginale in male Dermacentor andersoni transferred from parasitemic to susceptible cattle. Am J Vet Res. 1992;53:499–507.
Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. 2010;167:95–107.
Kocan KM, de la Fuente J, Blouin EF, Garcia-Garcia JC. Anaplasma marginale (Rickettsiales: Anaplasmataceae): recent advances in defining host-pathogen adaptations of a tick-borne rickettsia. Parasitology. 2004;129(Suppl):S285–300.
Kocan KM, de la Fuente J, Guglielmone AA, Meléndez RD. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev. 2003;16:698–712.
Radostits OM, Gay CC, Hinchcliff KW, Constable PD. Veterinary Medecine. A textbook of the diseaeses of cattle, horses, sheep, pigs and goats. 10th ed. New York: Elsevier Saunders; 2007.
Razmi GR, Dastjerdi K, Hossieni H, Naghibi A, Barati F, Aslani MR. An epidemiological study on Anaplasma infection in cattle, sheep, and goats in Mashhad Suburb, Khorasan Province, Iran. Ann N Y Acad Sci. 2006;1078:479–81.
de la Fuente J, Lew A, Lutz H, Meli ML, Hofmann-Lehmann R, Shkap V, et al. Genetic diversity of Anaplasma species major surface proteins and implications for anaplasmosis serodiagnosis and vaccine development. Anim Health Res Rev. 2005;6:75–89.
Ait Lbacha H, Alali S, Zouagui Z, Mamoun L El, Rhalem A, Petit E, et al. High Prevalence of Anaplasma spp. in Small Ruminants in Morocco. Transbound Emerg Dis. 2015. doi: 10.1111/tbed.12366.
Renneker S, Abdo J, Bakheit MA, Kullmann B, Beyer D, Ahmed J, et al. Coinfection of Sheep with Anaplasma, Theileria and Babesia Species in the Kurdistan Region, Iraq. Transbound Emerg Dis. 2013;60:113–8.
Renneker S, Abdo J, Salih DEA, Karagenç T, Bilgiç H, Torina A, et al. Can anaplasma ovis in small ruminants be neglected any longer? Transbound Emerg Dis. 2013;60:105–12.
Friedhoff KT. Tick-borne diseases of sheep and goats caused by Babesia, Theileria or Anaplasma spp. Parassitologia. 1997;39:99–109.
Manickam R. Epidemiological and clinical observations of acute anaplasmosis in sheep. Indian J Vet Med. 1987;7:159–60.
Noaman V, Shayan P. Molecular detection of Anaplasma bovis in cattle from central part of Iran. Vet Res Forum [Internet]; 2012;1:117–22. Available from: http://vrf.iranjournals.ir/article_1492_1.html.
Gaff HD, Kocan KM, Sonenshine DE. Tick-borne rickettsioses II (Anaplasmataceae). In: Sonenshine DE, Roe RM, editors. Biology of Ticks. Vol 2, Second ed. New York: Oxford University; 2014. p. 251–77.
Gordon WS, Brownlee A, Wilson DR, Macleod J. Tick-borne fever: a hitherto undescribed disease of sheep. J Comp Pathol Ther. 1932;45:301–7.
Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–95.
Gribble DH. Equine ehrlichiosis. J Am Vet Med Assoc. 1969;155:462–9.
Madewell BR, Gribble DH. Infection in two dogs with an agent resembling Ehrlichia equi. J Am Vet Med Assoc. 1982;180:512–4.
Bjöersdorff A, Svendenius L, Owens JH, Massung RF. Feline granulocytic ehrlichiosis-a report of a new clinical entity and characterisation of the infectious agent. J Small Anim Pract. 1999;40:20–4.
Chochlakis D, Ioannou I, Tselentis Y, Psaroulaki A. Human anaplasmosis and Anaplasma ovis variant. Emerg Infect Dis. 2010;16:1031–2.
Chochlakis D, Koliou M, Ioannou I, Tselentis Y, Psaroulaki A. Kawasaki disease and Anaplasma sp. infection of an infant in Cyprus. Int J Infect Dis. 2009;13:e71–3.
Bastos ADS, Mohammed OB, Bennett NC, Petevinos C, Alagaili AN. Molecular detection of novel Anaplasmataceae closely related to Anaplasma platys and Ehrlichia canis in the dromedary camel (Camelus dromedarius). Vet Microbiol. 2015;179:310–4.
Belkahia H, Said MB, Sayahi L, Alberti A, Messadi L. Detection of novel strains genetically related to Anaplasma platys in Tunisian one-humped camels (Camelus dromedarius). J Infect Dev Ctries. 2015;9:1117–25.
Li Y, Yang J, Chen Z, Qin G, Li Y, Li Q, et al. Anaplasma infection of Bactrian camels (Camelus bactrianus) and ticks in Xinjiang, China. Parasit Vectors [Internet]. 2015;8,313:1-6. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4464610/.
Ben Said M, Belkahia H, Sayahi L, Aloui M, Jemli MH, Hadj Mohamed B, et al. [First serological study of the prevalence of Anaplasma phagocytophilum in dromedary (Camelus dromedarius) in Tunisia]. Bull Soc Pathol Exot. 2014;107:1–6. 1990.
Walker AR, Bouattour A, Camicas JL, Estrada-Pena A, Horak IG, Latif AA, et al. Ticks of domestic animals in Africa: a guide to identification of species. Bioscience Reports. Edinburgh: University of Edinburgh; 2003. p. 221.
Ybañez AP, Matsumoto K, Kishimoto T, Inokuma H. Molecular analyses of a potentially novel Anaplasma species closely related to Anaplasma phagocytophilum detected in sika deer (Cervus nippon yesoensis) in Japan. Vet Microbiol. 2012;157:232–6.
Drazenovich N, Foley J, Brown RN. Use of real-time quantitative PCR targeting the msp2 protein gene to identify cryptic anaplasma phagocytophilum infections in wildlife and domestic animals. Vector Borne Zoonotic Dis. 2006;6:83–90.
Abramson JH. WINPEPI updated: computer programs for epidemiologists, and their teaching potential. Epidemiol Perspect Innov. 2011;8:1.
Atarhouch T, Rami M, Bendahman MN, Dakkak A. Camel trypanosomosis in Morocco 1: results of a first epidemiological survey. Vet Parasitol. 2003;111:277–86.
Rami M, Atarhouch T, Bendahman MN, Azlaf R, Kechna R, Dakkak A. Camel trypanosomosis in Morocco: 2. A pilot disease control trial. Vet Parasitol. 2003;115:223–31.
Sudan V, Sharma RL, Borah MK. Subclinical anaplasmosis in camel (Camelus dromedarius) and its successful therapeutic management. J Parasit Dis. 2014;38:163–5.
Ybañez AP, Matsumoto K, Kishimoto T, Yokoyama N, Inokuma H. Dual presence of anaplasma phagocytophilum and its closely related anaplasma sp. In ixodid ticks in Hokkaido, Japan, and their specific molecular detection. J Vet Med Sci. 2012;74:1551–60.
Ybañez AP, Tagawa M, Matsumoto K, Kishimoto T, Yokoyama N, Inokuma H. Specific molecular detection of Anaplasma sp. Closely related to Anaplasma phagocytophilum in ixodid ticks and cattle in a pastureland in Hokkaido, Japan. Vector Borne Zoonotic Dis. 2013;13:6–11.
Al-Adhami B, Scandrett WB, Lobanov VA, Gajadhar AA. Serological cross-reactivity between Anaplasma marginale and an Ehrlichia species in naturally and experimentally infected cattle. J Vet Diagn Invest. 2011;23:1181–8.
Dreher UM, de la Fuente J, Hofmann-Lehmann R, Meli ML, Pusterla N, Kocan KM, et al. Serologic cross-reactivity between Anaplasma marginale and Anaplasma phagocytophilum. Clin Diagn Lab Immunol. 2005;12:1177–83.
Mentaberre G, Gutiérrez C, Rodríguez NF, Joseph S, González-Barrio D, Cabezón O, et al. A transversal study on antibodies against selected pathogens in dromedary camels in the Canary Islands, Spain. Vet Microbiol. 2013;167:468–73.
Ajayi SA, Onyali IO, Oluigbo FO, Ajayi ST. Serological evidence of exposure to Anaplasma marginale in Nigerian one-humped camels. Vet Rec. 1984;114:478.
Bamaiyi PH, Kalu AU, Ali M. Heamoparasites of the trade camel (Camelus dromedarius) slaughtred in Maiduguri, Borno State, Nigeria. Cont J Vet Sci. 2011;5:18–21.
Mohammed A, Sackey A, Tekdek L, Gefu J. Common health problems of the one humped camel (Camelus dromedarius) introduced into sub-humid climate in Zaria, Nigeria. Res J Anim Sci. 2007;1:1–5.
Rabana JL, Kumshe HA, Kamani J, Hafsat G, Turaki UA, Dilli HK. Effects of parasitic infections on Erythrocyte indices of camels in Nigeria. Vet Res Forum. 2012;2:59–63.
Demoncheaux J-P, Socolovschi C, Davoust B, Haddad S, Raoult D, Parola P. First detection of Rickettsia aeschlimannii in Hyalomma dromedarii ticks from Tunisia. Ticks Tick-Borne Dis. 2012;3:398–402.
Djerbouh A, Kernif T, Beneldjouzi A, Socolovschi C, Kechemir N, Parola P, et al. The first molecular detection of Rickettsia aeschlimannii in the ticks of camels from southern Algeria. Ticks Tick-Borne Dis. 2012;3:374–6.
Kernif T, Djerbouh A, Mediannikov O, Ayach B, Rolain J-M, Raoult D, et al. Rickettsia africae in Hyalomma dromedarii ticks from sub-Saharan Algeria. Ticks Tick-Borne Dis. 2012;3:377–9.
Sarih M, M’Ghirbi Y, Bouattour A, Gern L, Baranton G, Postic D. Detection and identification of ehrlichia spp. In ticks collected in Tunisia and morocco. J Clin Microbiol. 2005;43:1127–32.
Bailly-Choumara H, Morel P, Rageau J. Sommaire des données actuelles sur les tiques du Maroc (Acari, Ixodidea). Bull Inst Sci. 1976;1:101–7.
Gharbi M, Moussi N, Jedidi M, Mhadhbi M, Sassi L, Darghouth MA. Population dynamics of ticks infesting the one-humped camel (Camelus dromedarius) in central Tunisia. Ticks Tick-Borne Dis. 2013;4:488–91.
Gharbi M, Aziz Darghouth M. A review of Hyalomma scupense (Acari, Ixodidae) in the Maghreb region: from biology to control. Parasite [Internet]. 2014;21(2):1-12. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917376/.
Acknowledgements
We thank the veterinarians, laboratory technicians, farmer cooperative members and livestock keepers who helped with this work. We thank Prof Ian Maudlin of the University of Edinburgh for his careful reading of the manuscript.
Funding
This work was supported by the PRAD project under grant agreement number 28027YM, the Institut Agronomique et Vétérinaire Hassan II, BIPAR and Ecole Nationale Vétérinaire de Toulouse.
Availability of data and materials
Not applicable.
Authors’ contributions
HAL, ZZ, SA, AR, HJB, RM designed and coordinated the work. HAL and EP performed the laboratory analysis under the supervision of HJB and RM. HAL and RM analysed the data and wrote the manuscript. MJD translated revised and edited the manuscript. All authors read, commented and approved the final manuscript.
Authors’ information
HAL is a lecturer in ruminant medicine and surgery and a doctoral researcher at the Institut Agronomique et Veterinaire Hassan II (IAVHII) who has been working on anaplasmosis in ruminants and camelids for the last three years. ZZ and SA are professors in ruminant medicine and surgery and researchers at the IAVHII. AR is a professor and researcher in parasitology at the IAVHII. MJD is a researcher in neglected zoonoses previously based at the University of Edinburgh and now working for Ceva Sante Animale. EP is an assisting engineer and responsible for PCR analysis and supervision of sequencing in the research unit involved in vectorized bacteria within BIPAR. HJB is the head of the research unit involved in vectorized bacteria within BIPAR and responsible for the Bartonella and Anaplasma research within BIPAR. RM is an assistant professor in cattle medicine and co-responsible for the PRAD research project between Morocco and France dedicated to Anaplasma in ruminants and camelids.
Competing interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Ethics approval
Approval for the study went through the official channels of the Institut Agronomique et vétérinaire Hassan II (Maroc), Moroccan Ministry of Agriculture and Marine Fisheries. As veterinarians at the IAV Hassan, we are also bound by the application of ordinal Moroccan law relating to respect of ethics and animal welfare (Décret n°2-07-1332 du 5 rabii II 1431 (22 mars 2010)) in keeping with EU ethical standards. Study participants were briefed on the purpose of the study and informed consent was therefore obtained.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Multilingual abstracts in the six official working languages of the United Nations. (PDF 722 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ait Lbacha, H., Zouagui, Z., Alali, S. et al. “Candidatus anaplasma camelii” in one-humped camels (Camelus dromedarius) in Morocco: a novel and emerging Anaplasma species?. Infect Dis Poverty 6, 1 (2017). https://doi.org/10.1186/s40249-016-0216-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40249-016-0216-8