Abstract
A molecular-imprinted polymer nanoparticles (MIP-NP) for the selective preconcentration of 2,4-dichlorophenoxyacetic acid (2,4-D) is described. It was obtained by precipitation polymerization from methacrylic acid (the functional monomer), ethylene glycol dimethacrylate (the cross-linker), 2,2′-azobisisobutyronitrile (the initiator) and 2,4-D (the template molecule) in acetonitrile solution. The MIP-NPs were characterized by thermogravimetric analysis, and by scanning electron microscopy. Imprinted 2,4-D molecules were removed from the polymeric structure using acetic acid in methanol (15:85 v/v %) as the eluting solvent. The sorption and desorption process occur within 10 min and 15 min, respectively. The maximum sorbent capacity of the molecular imprinted polymer is 89.2 mg g−1. The relative standard deviation and limit of detection for water samples by introduced selective solid phase extraction were 4.2% and 1.25 μg L−1, and these data for urine samples were 4.7% and 1.80 μg L−1, respectively. The method was applied to the determination of 2,4-D in the urine and different water samples.
Similar content being viewed by others
Introduction
In recent years, the hazards of using pesticides have been accentuated by the sharp rise in their use in agriculture and industry. Phenoxy herbicides compounds are currently among the most frequently used pesticides worldwide [1]. They have been used on a large scale in agriculture to control the growth of broad-leaved weeds on rice, maize, wheat, and in post-emergence applications in most developing countries [2],[3]. Among them, 2,4-dichlorophenoxyacetic acid (2,4-D) is a common important phenoxy herbicide that is selective, systemic auxin-type herbicide extensively used throughout the world for the past 50 years [4]. Exposures to 2,4-D occurs not only in general population groups through trace residues in the diet, from drinking water and direct contact with lawns recently treated with 2,4-D [5] but also persons who occupationally handle (mix, load or apply) it. 2,4-D is rapidly absorbed into the body following ingestion, inhalation and/or dermal contact and is rapidly excreted, predominantly in the urine unchanged [6],[7]. Some recent well-documented studies show an association between the agricultural use of the 2,4-dichlorophenoxyacetic acid (2,4-D) and the risk of non- Hodgkin’s lymphoma [8]. It has been classified as possibly carcinogenic to humans (IARC, group 2B) [4]. Furthermore, continuous use of 2,4-D may cause soil percolation and groundwater contamination [9]. Due to the presence of the carboxylate group in its structure, this compound can react with metal ions to form complexes which are sparingly soluble in water. The World Health Organization (WHO), U.S. Environmental Protection Agency (EPA), and national governments such as New Zealand and United State of America (USA) regulation set the maximum contaminant level (MCL) of chlorophenoxy acid herbicides in drinking water in the range of 10-70 ng mL−1[10],[11]. Because of its adverse effect on humans, sensitive determination of 2,4-D in environmental and biological samples is necessary.
In the literature, several methods have been developed for determining 2,4-D in different environmental samples, including solid phase microextraction gas chromatography-mass spectrometry (SPME-GC-MS) [12], solid phase extraction high-performance liquid chromatographic with UV detector (SPE-HPLC-UV) [13] and liquid - liquid extraction gas chromatography-mass spectrometry (LLE-GC-MASS) [14] methods. The mentioned methods have major drawbacks as they are very time consuming procedure, requiring expensive instrumentation, affected by matrix effects and have low selectivity and recoveries. According to the complexity of matrix and low content of 2,4-D in real samples, an efficient sample treatment technique with high clean-up and high enrichment factor are required prior to analysis. Thus, development of a rapid and selective extraction technique for determination of 2,4-D from environmental and biological matrices is of great importance.
In recent years, molecularly imprinted polymers (MIPs) and ion imprinted polymers (IIPs) have been extensively used (e.g. SPE sorbent) for preconcentration and high efficient separation of different trace analytes in complex matrices [15]-[28], based on their unique imprinting and recognition properties. MIPs demonstrate high potential for sample clean-up of very complicated matrix [29], solid-phase extraction [30]-[34], and solid-phase micro extraction fibers [35]. The most widely employed preparation method for synthesis of MIPs is bulk polymerization. The prepared polymer with this method is then ground and sieved, and particles of usually about 25 mm are collected for subsequent studies.
Although the bulk MIP prepared by conventional methods exhibits high selectivity [36],[37], some disadvantages were suffered, such as the heterogeneous distribution of the binding sites, embedding of most binding sites, and poor site accessibility for template molecule [38]. Consequently, the consideration of researchers has moved toward achieving highly uniform spherical imprinted particles, particularly on the nanoscale [39]. In contrast to monoliths, MIPs possess cavities designed for a target analyte, providing a retention mechanism based on molecular recognition; by this, imprinted cavities are more easily available to templates and the binding kinetic is increased [40]. Different methods are used to obtain MIPs Nano particles, namely suspension, multistep swelling and precipitation polymerization. The different methods of preparation have been well documented in a recent review by Haginaka [41]. Among them, the precipitation technique is the most convenient one, because it is a homogeneous, one-step synthesis and does not require the use of a surfactant or stabilizer that can remain adsorbed to the surface of the polymer, interfering with the selective binding of the target to the imprinted materials. The first prepared MIPs, using precipitation polymerization technique, were reported by Ye et al. [42]. The polymer particles were obtained according to this procedure, have more uniform particles size and require no crushing and sieving steps in comparison tothe polymers obtained by bulk polymerization.
The aim of the present study was the preparation of 2,4-D imprinting polymer via non-covalent precipitation polymerization as well as its application in selective sample preparation in order to eliminate the matrix effects. HPLC-UV was used for determination of 2,4-D after preconcentration by MIP-NPs in urine and different water samples.
Experimental
Reagents and chemicals
2,4-D was purchased from Fluka (Buchs, Switzerland). Azobisisobutyronitrile (AIBN) as the initiator was obtained from Acros Organic (NJ, USA). Methacrylic acid (MAA), ethylene glycol dimethacrylare (EGDMA), acetic acid (HOAC), sodium hydroxide (NaOH), acetonitrile (ACN) and methanol (MeOH) were purchased from Merck (Darmstadt, Germany, www.merck.de). All the reagents used were of analytical grade. Ultrapure water was prepared using a Milli-Q® system (Millipore, Milford, MA, USA). Stock standard solution of 2,4-D (1000 mg L−1) was prepared in methanol and stored at refrigerator.
Instrumentation
All measurements were performed by a reversed-phase HPLC system from the Knauer Company (Germany), consisting of a K-1001 series high-pressure pump, a K-2006 photo diode-array detector and a VS injection valve, equipped with a 20-μL loop. The analytes were separated on a Chromolith performance RR-C18 100 mm × 4.6 mm i.d. (MerchKGa A, Germany) and column guards (Chromolith Guard Cartridge Kit RP- C18 and 5 cm × 4.6 mm i.d., 5 μm). The mobile phase composition was 79.5% acetonitrile, 19.5% purified water and 1% acetic acid The flow-rate was set at 1 mL min−1 in isocratic mode with the detector’s wavelength set at 280 nm. A digital pH meter, WTW Metrohm 827 Ion analyzer (Herisau, Switzerland), equipped with a combined glass calomel electrode was used for the pH adjustments at 25 ± 1°C temperature. Scanning electron microscopy (SEM) was carried out by gently distributing the powder sample on the stainless steel stubs, using SEM (Philips, XL-30, Almelo, the Netherlands) instrument. BAHR-Thermoanalyse GmbH (Germany) was used to determine the thermal properties of synthesized polymers with employing, heating and cooling rates of 10 C°min−1 and using a condenser as the coolant. The samples were weighed as a thin film and carefully packed into a clean aluminum pan (11.5-12.5 mg), and sealed by crimping an aluminum lid on the pan (Shimadzu universal crimper). An Al2O3 empty pan sealed with a cover pan was used as a reference sample. A scanning range of 10 to 800°C was used for samples at 10°C min−1 in nitrogen gas.
Synthesis of molecular imprinted and non imprinted polymer nanoparticles
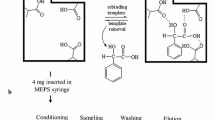
Precipitation polymerization technique was used for preparation of MIP-NPsaccording to following procedure (Figure 1): 1 mmol of 2,4-D (template) 4 mmol of MAA (functional monomer)which were dissolved in 40 mL of acetonitrile (progen solvent)in a 100 mL glass flask. The solution was mixed by ultrasonic wave for 10 min and then 1 mmol of AIBN (initiator) and 25 mmol of EGDMA (cross-linker agent) were added to the solution. The polymerization mixture was purged with a gentle flow of nitrogen gas for 6 min and the flask was sealed under nitrogen then; polymerization was performed in an oil bath at 60°C for 24 h in the presence of nitrogen under magnetic stirring at 400 rpm. The polymer nanoparticles were collected by centrifugation and washed with acetone and methanol, respectively for removing additional reagents and solvent. The template was removed from the cavity of MIP-NP using soxhlet extraction (acetic acid-methanol (85:15 v/v)), This step was confirmed through analysis of the eluted template molecules by HPLC analysis. Afterwards, the polymer was washed with deionized water until neutralization was obtained. Furthermore, non-imprinted polymers (NIP), as a reference, was prepared using the same process without the addition of the template molecule.
Preparation of standard solutions and real samples
Stock standard solution of the 2,4-D was prepared at concentration level of 1000 mg L−1 in methanol. The stock solution was protected from light and stored at 4°C and brought to ambient temperature just prior to use. The required working solutions were prepared daily in double-distilled water prior to use. For analysis of urine sample, 2 mL of spiked urine was diluted to 7.0 mL with ultrapure water. The water samples, including distilled water, tap water (Tehran, Iran), river waters obtained from Siahrood river and Derka river (Ghaemshahr, Iran) and sea water (Caspian Sea, Sari, Iran) were collected in cleaned polyethylene bottles and filtered through a 0.45 mm pore size nylon filter (Millipore) immediately after sampling. The amounts of 2,4-D was successfully determined by the present method.
Extraction procedure
All factors affecting sorption and desorption steps were investigated using batch method. Extraction of 2,4-D molecules from modeling solutions and real samples is followed by two steps: sorption and desorption. In the sorption step, the pH of sample solution was adjusted to 4.0 by drop wise addition of 2 mol L−1 sodium hydroxide or hydrochloric acid solutions. Then, 50 mg of dried polymer was suspended in aqueous solution (10 mL) containing 0.02 mg L−1 concentration of 2,4-D and stirred for 10 min with a magnetic stirrer, after that the polymer nanoparticles were separated from the solution by centrifugation. In washing step, in order to remove the potential interfering compounds and loosely retained molecules, the MIP-NPs were washed several times with double distilled water, then in desorption step, the elution of 2,4-D from MIP-NPs was performed by 1.5 mL of HOAC in methanol (15:85 v/v %) for 15 min. The eluate (separated by centrifuge) evaporated to almost dry under a gentle stream of nitrogen and the residue was dissolved in 0.3 mL mobile phase under sonication. Finally, 20 μL of the separated phase was drawn out by a Hamilton syringe and directly injected into the HPLC-Uv for analysis. Figure 2 provides a scheme for illustration of the mentioned extraction procedure.
Extraction percent of 2,4-D was calculated by the following equation:
CA and CB are the concentrations of 2,4-D before and after extraction in the solution, respectively.
Results and discussion
Characterization of the synthesized molecular imprinted polymer nanoparticles
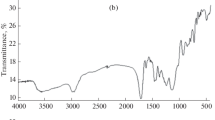
The resulting nanosized imprinted polymers were characterized by scanning electron microscopy (SEM), and thermogravimetric analysis (TGA). Thermal stability of the synthesized imprinted polymer nanoparticles was evaluated by TGA. Figure 3 shows TGA plot for the imprinted polymer. TGA plot for MIP-NPs can prove the thermal stability of the synthesized polymer. The morphology of the MIP-NPs was assessed by scanning electron microscopy, and the SEM micrograph is shown in Figure 4. As you can see in the Figure 4, the particle size of synthesized imprinted polymer was in range of 25-37 nm. These observation indicate that the formation 2,4-D imprinted nano particles was performed successfully and it can be used as a nano-size selective solid phase for very fast extraction of ultra-trace amounts of 2,4-D. The prepared MIP in the nanosize particles have many advantages such as: fast extraction of analyte from the real samples, high adsorption capacity of the prepared nanosized MIP toward conventional MIP according to the simple diffusion of target molecules to formed cavity due to high surface area of nanosized MIP.
Removal step
Effect of solution’s pH
To evaluate the effect of pH on the extraction efficiency, the pH of 10 mL of sample solutions containing 0.02 mg L−1 of 2,4-D was adjusted in the range of 2-9. The pH of the suspensions was adjusted to desired values by adding sodium hydroxide or hydrochloric acid. Figure 5 illustrates the effect of solution’s pH on percent of Adsorption efficiency. As it shows, the adsorption of 2,4-D by MIP-NPs was approximately constant from pH 2.0 to 4.0 but decreased from pH 5.0 to 9.0. Therefore, the pH of 4.0 was chosen as optimized pH, for quantitative adsorption of 2,4-D on the synthesized MIP-NPs in the subsequent experiments.
Equilibrium sorption time
In a typical uptake kinetics test, 50 mg of the sorbent was added to 10 mL of 0.02 mg L−1 2,4-D aqueous solution at pH 4.0. The resulting suspension was stirred in different times (i.e., from 5 to 20 min) under magnetic stirring (Figure 6a). Consequently, an optimum equilibration time of 10 minutes was obtained for quantitative removal of 2,4-D from solution into the solid phase.
Desorption step
Choice of eluent and desorption time
Complete removal of template from synthesized polymer isa very crucial step to guarantee the absence of memory effect. In this step, several solvents were examined; whose results are provided in Table 1. It is obvious that HOAC in methanol (15:85 v/v %) shows better recovery compared with the other solvents. Acetic acid can affect the hydrogen bonding between template and functional monomer, so that the removal of the template would be easier. In subsequent experiments (Table 2), 1.5 mL of HOAC in methanol (15:85 v/v %) was used as optimum eluent.
In order to investigate the optimum desorption time, various times were examined in the range of 5 to 25 min, while other parameters were kept in optimum conditions. According to measurements, extraction recovery was increased up to 15 min and it was constant in longer times. Therefore, 15 minutes can be considered to be the best quantitative time for the elution of 2,4-D from the imprinted polymer (Figure 6b).
The effect of centrifugation time
The final parameter which evaluated and optimized was centrifugation time. If the centrifugation time is not enough, the MIP nanoparticles cannot be completely collected on bottom of the vial. A series of extraction with varying centrifugation times from 1 to 5 min at a rate of 3000 rpm were performed. The extraction recovery for the analytes was lower when the centrifugation time was shorter than 3 min. However; longer centrifugation has not significant effect on the extraction efficiency of the target molecule. Therefore, 3 min was selected as the optimum centrifuging time.
Investigation of the selectivity of the synthesized molecular imprinted polymer for 2,4-D
The effect of interfering agents on extraction recovery of 2,4-D were investigated in the presence of methylbenzene, aniline and 2-(4-chloro-2-methylphenoxy) propanoic acid with hundred fold concentration as interfering agents separately, while the other parameters were considered to be constant. The extraction recovery of more than 95% was obtained for 2-4-D in the presence of potentially interfering agent because of the high selectivity of synthesized MIP-NPs that allows its potential application as a sample preparation and clean up technique for analysis of 2,4-D in different matrices in the presence of interfering agents.
Comparison of MIP and NIP
In order to compare MIP and NIP, two extractions by both MIP and NIP in water sample were assessed under the optimal conditions. As it has shown (Table 3), because of the high capacity of MIP for extraction of 2,4-D in comparison with NIP, the extraction recovery of MIP is more than that of NIP. The obtained results can prove the successful engineering of cavity for 2,4-D in the structure of synthesized MIP-NPs. In order to evaluate the maximum adsorption capacity, the difference between concentration of the solution before extraction and the concentration of the solution after extraction was calculated. The sorption capacities of the MIP and NIP were calculated to be 89.2 mg g−1 and 12.1 mg g−1, respectively.
The effect of interfering agent
The effect of interfering agents on the extraction recovery of 2,4-D were investigated in the presence of atrazine, 3,4-Dichlorophenol and 2,4,6-Trichlorophenol with tenfold concentration as interfering agents separately, while, the other parameters were considered to be constant. The extraction recovery of more than 95% was obtained for 2,4-D in the presence of potentially interfering agents because of the high selectivity of MIP that allows its potential application as a sample preparation tool for analysis of 2,4-D extracted from environmental and biological samples.
Statistical and calibration parameters
Under optimized conditions that have been described, the MIP-NPs-SPE showed a linear calibration curve within concentration ranging from 5 to 800 μg L−1. The least square equations at above linear dynamic range for water and urine samples were as follows:
The limits of detections, defined as CLOD =3Sb/m, where Sb is the standard deviation of seven replicate blank signals and m is the slope of the linear section of calibration curve after preconcentration, for a sample volume of 10 mL, were found to be 1.25 μg L−1 and 1.80 μg L−1 for 2,4-D in water and urine samples, respectively. Also, the relative standard deviations for eight separate batch experiments with 50 mg of sorbent for determination of 0.1 μg 2,4-D in 10 mL of water were calculated to be 4.2% and 4.7% in water and urine samples, respectively.
Determination of 2,4-D in different water and urine samples
In order to evaluate the analytical applicability of the proposed method, determination of 2,4-D in urine and different water samples such as distilled water, tap water (Tehran, Iran), river waters obtained from Siahrood river and Derka river (Ghaemshahr, Iran) and sea water (Caspian Sea, Sari, Iran) was carried out. Table 4 shows that the results of three replicate analyses of each real sample obtained by the method are in good agreement with the spiking amounts. The repeatability of the method was demonstrated by the mean relative standard deviation (R.S.D.). Also, the relative recovery is defined as following equation:
Where Cfound, Creal, and Cadded are the concentrations of analyte after addition of known amount of standard in the real sample, the concentration of analyte in real sample and the concentration of known amount of standard which was spiked to the real sample, respectively. 2,4-D trace determination in water and urine samples, using MIP-NPs-SPE as a reliable sample treatment technique can be conducted. Figure 7 provides chromatograms for determination of 2,4-D in spiked urine and sea water samples.
Conclusion
The molecular imprinted polymer is a selective sample preparation technique for preconcentration of molecules such as 2,4-D from aqueous solutions. The MIP-NPs has been synthesized by precipitation polymerization technique. The analytical characteristics such as good precision and preconcentration factor, high quantities of recoveries, wide dynamic linear range, and low detection of limit were achieved due to the powerful efficiency of the MIP-NPs-SPE method. Other advantages of suggested method are: lack of matrix effect, low consumption of organic solvent, environmentally friendly, safe, simplicity and selectivity. The synthesized MIP-NPs can be used repeatedly (7 times) with no significant decrease in its binding affinities. Due to relatively high preconcentration factor, trace amount of 2,4-D at μg L−1 levels can be determined and separated by MIP-NPs.
Authors’ contributions
The overall implementation of this study including design, experiments and data analysis, and manuscript preparation were the results of efforts by all of the authors. All authors have made extensive contribution into the review and finalization of this manuscript. All authors read and approved the final manuscript.
References
Garabrant DH, Philbert MA: Review of 2, 4-dichlorophenoxyacetic acid (2, 4-D) epidemiology and toxicology. CRC Crit Rev Toxicol 2002, 32(4):233–257. 10.1080/20024091064237
Chaudhary AJ, M-u H, Grimes SM: Simultaneous recovery of metals and degradation of organic species: copper and 2, 4, 5-trichlorophenoxyacetic acid (2, 4, 5-T). J Hazard Mater 2009, 165(1):825–831. 10.1016/j.jhazmat.2008.10.066
de Lipthay JR, Sørensen SR, Aamand J: Effect of herbicide concentration and organic and inorganic nutrient amendment on the mineralization of mecoprop, 2, 4-D and 2, 4, 5-T in soil and aquifer samples. Environ Pollut 2007, 148(1):83–93. 10.1016/j.envpol.2006.11.005
IARC: IARC monographs on the evaluation of carcinogenic risksto humans: chlorophenoxy herbicides. 1998, available on line ., [http://www-cie.iarc.fr]
Reregistration Eligibility Decision for 2,4-D. EPA 738-R-05–002. 2005.
Kohli J, Khanna R, Gupta B, Dhar M, Tandon J, Sircar K: Absorption and excretion of 2, 4-dichlorophenoxyacetic acid in man. Xenobiotica 1974, 4(2):97–100. 10.3109/00498257409049349
Sauerhoff M, Braun W, Blau G, Gehring P: The fate of 2, 4-dichlorophenoxyacetic acid (2, 4-D) following oral administration to man. Toxicology 1977, 8(1):3–11. 10.1016/0300-483X(77)90018-X
Toxicology of the Blood and Bone Marrow. Raven Press, New York; 1985.
Harris S, Solomon K, Stephenson G: Exposure of homeowners and bystanders to 2, 4-dichlorophenoxyacetic acid (2, 4-D). J Environ Sci Health B 1992, 27(1):23–38. 10.1080/03601239209372765
National Primary Drinking Water Regulations, Office of Water, U.S. Environmental Protection Agency, Washington, DC.; 2009.
Hamilton D, Ambrus A, Dieterle R, Felsot A, Harris C, Holland P, Katayama A, Kurihara N, Linders J, Unsworth J, Wong S: Regulatory limits for pesticide residues in water (IUPAC Technical Report). Pure Appl Chem 2003, 75(8):1123–1155. 10.1351/pac200375081123
Rodríguez I, Rubí E, González R, Quintana JB, Cela R: On-fibre silylation following solid-phase microextraction for the determination of acidic herbicides in water samples by gas chromatography. Anal Chim Acta 2005, 537(1–2):259–266. 10.1016/j.aca.2004.12.083
Biesaga M, Jankowska A, Pyrzyńska K: Comparison of different sorbents for solid-phase extraction of phenoxyalkanoic acid herbicides. Microchim Acta 2005, 150(3–4):317–322. 10.1007/s00604-005-0359-y
Catalina MI, Dallüge J, Vreuls RJ, Brinkman UAT: Determination of chlorophenoxy acid herbicides in water by in situ esterification followed by in-vial liquid–liquid extraction combined with large-volume on-column injection and gas chromatography–mass spectrometry. J Chromatogr A 2000, 877(1):153–166. 10.1016/S0021-9673(00)00242-9
Jing T, Wang Y, Dai Q, Xia H, Niu J, Hao Q, Mei S, Zhou Y: Preparation of mixed-templates molecularly imprinted polymers and investigation of the recognition ability for tetracycline antibiotics. Biosens Bioelectron 2010, 25(10):2218–2224. 10.1016/j.bios.2010.02.023
Behbahani M, Salarian M, Bagheri A, Tabani H, Omidi F, Fakhari A: Synthesis, characterization and analytical application of Zn (II)-imprinted polymer as an efficient solid-phase extraction technique for trace determination of zinc ions in food samples. J Food Compos Anal 2014, 34: 81–89. 10.1016/j.jfca.2013.10.003
Hoshina K, Horiyama S, Matsunaga H, Haginaka J: Molecularly imprinted polymers for simultaneous determination of antiepileptics in river water samples by liquid chromatography–tandem mass spectrometry. J Chromatogr A 2009, 1216(25):4957–4962. 10.1016/j.chroma.2009.04.071
Behbahani M, Barati M, Bojdi MK, Pourali AR, Bagheri A, Tapeh NAG: A nanosized cadmium (II)-imprinted polymer for use in selective trace determination of cadmium in complex matrices. Microchim Acta 2013, 180(11–12):1117–1125. 10.1007/s00604-013-1036-1
Prieto A, Schrader S, Bauer C, Möder M: Synthesis of a molecularly imprinted polymer and its application for microextraction by packed sorbent for the determination of fluoroquinolone related compounds in water. Anal Chim Acta 2011, 685(2):146–152. 10.1016/j.aca.2010.11.038
Lucci P, Núñez O, Galceran MT: Solid-phase extraction using molecularly imprinted polymer for selective extraction of natural and synthetic estrogens from aqueous samples. J Chromatogr A 2011, 1218(30):4828–4833. 10.1016/j.chroma.2011.02.007
Behbahani M, Bagheri S, Amini MM, Sadeghi Abandansari H, Moazami HR, Bagheri A: Application of a magnetic molecularly imprinted polymer for the selective extraction and trace detection of lamotrigine in urine and plasma samples. J Sep Sci 2014, 37: 1610–1616. 10.1002/jssc.201400188
Shahtaheri S, Katmeh M, Kwasowski P, Stevenson D: Development and optimisation of an immunoaffinity-based solid-phase extraction for chlortoluron. J Chromatogr A 1995, 697(1):131–136. 10.1016/0021-9673(94)01088-V
Koohpaei A, Shahtaheri S, Ganjali M, Forushani AR, Golbabaei F: Application of multivariate analysis to the screening of molecularly imprinted polymers (MIPs) for ametryn. Talanta 2008, 75(4):978–986. 10.1016/j.talanta.2007.12.046
Koohpaei A, Shahtaheri S, Ganjali M, Forushani AR, Golbabaei F: Optimization of solid-phase extraction using developed modern sorbent for trace determination of ametryn in environmental matrices. J Hazard Mater 2009, 170(2):1247–1255. 10.1016/j.jhazmat.2009.05.125
Bulut VN, Gundogdu A, Duran C, Senturk HB, Soylak M, Elci L, Tufekci M: A multi-element solid-phase extraction method for trace metals determination in environmental samples on Amberlite XAD-2000. J Hazard Mater 2007, 146(1–2):155–163. 10.1016/j.jhazmat.2006.12.013
Soylak M, Ercan O: Selective separation and preconcentration of copper (II) in environmental samples by the solid phase extraction on multi-walled carbon nanotubes. J Hazard Mater 2009, 168(2–3):1527–1531. 10.1016/j.jhazmat.2009.03.032
Ebrahimzadeh H, Behbahani M: A novel lead imprinted polymer as the selective solid phase for extraction and trace detection of lead ions by flame atomic absorption spectrophotometry: synthesis, characterization and analytical application. Arab J CHEM 2013, doi:10.1016/j.arabjc.2013.09.017.
Omidi F, Behbahani M, Samadi S, Sedighi A, Shahtaheri SJ: Coupling of molecular imprinted polymer nanoparticles by high performance liquid chromatography as an efficient technique for sensitive and selective trace determination of 4-chloro-2-methylphenoxy acetic acid in complex matrices. Iranian J Publ Health 2014, 43(5):645–657.
Wang T, Tong J, Sun M, Chen L: Fast and selective extraction of chloramphenicol from soil by matrix solid-phase dispersion using molecularly imprinted polymer as dispersant. J Sep Sci 2011, 34(15):1886–1892. 10.1002/jssc.201100046
Tamayo FG, Casillas JL, Martin-Esteban A: Highly selective fenuron-imprinted polymer with a homogeneous binding site distribution prepared by precipitation polymerisation and its application to the clean-up of fenuron in plant samples. Anal Chim Acta 2003, 482(2):165–173. 10.1016/S0003-2670(03)00213-7
Haginaka J, Sanbe H: Uniform-sized molecularly imprinted polymers for 2-arylpropionic acid derivatives selectively modified with hydrophilic external layer and their applications to direct serum injection analysis. Anal Chem 2000, 72(21):5206–5210. 10.1021/ac0005215
Turiel E, Martín-Esteban A, Tadeo JL: Molecular imprinting-based separation methods for selective analysis of fluoroquinolones in soils. J Chromatogr A 2007, 1172(2):97–104. 10.1016/j.chroma.2007.10.003
He J, Lv R, Zhu J, Lu K: Selective solid-phase extraction of dibutyl phthalate from soybean milk using molecular imprinted polymers. Anal Chim Acta 2010, 661(2):215–221. 10.1016/j.aca.2009.12.029
Yan H, Qiao J, Pei Y, Long T, Ding W, Xie K: Molecularly imprinted solid-phase extraction coupled to liquid chromatography for determination of Sudan dyes in preserved beancurds. Food Chem 2012, 132(1):649–654. 10.1016/j.foodchem.2011.10.105
Djozan D, Ebrahimi B: Preparation of new solid phase micro extraction fiber on the basis of atrazine-molecular imprinted polymer: application for GC and GC/MS screening of triazine herbicides in water, rice and onion. Anal Chim Acta 2008, 616(2):152–159. 10.1016/j.aca.2008.04.037
Akiyama T, Hishiya T, Asanuma H, Komiyama M: Molecular imprinting of cyclodextrin on silica-gel support for the stationary phase of high-performance-liquid-chromatography. J Inclusion Phenom Macrocyclic Chem 2001, 41(1–4):149–153. 10.1023/A:1014446420222
Zhang H, Dramou P, He H, Tan S, Pham-Huy C, Pan H: Molecularly imprinted stationary phase prepared by reverse micro-emulsion polymerization for selective recognition of gatifloxacin in aqueous media. J Chromatogr Sci 2012, 50(6):499–508. 10.1093/chromsci/bms028
Kan X, Zhao Y, Geng Z, Wang Z, Zhu J-J: Composites of multiwalled carbon nanotubes and molecularly imprinted polymers for dopamine recognition. J Phys Chem C 2008, 112(13):4849–4854. 10.1021/jp077445v
Ye L, Mosbach K, Spivak DA, Sibrian-Vazquez M: Molecularly Imprinted Materials: Towards the Next Generation. Materials Research Society Symposium Proceedings. Cambridge Univ Press; 2002.
Tokonami S, Shiigi H, Nagaoka T: Review: micro- and nanosized molecularly imprinted polymers for high-throughput analytical applications. Anal Chim Acta 2009, 641(1–2):7–13. 10.1016/j.aca.2009.03.035
Haginaka J: Monodispersed, molecularly imprinted polymers as affinity-based chromatography media. J Chromatogr B 2008, 866(1–2):3–13. 10.1016/j.jchromb.2007.07.019
Ye L, Cormack PA, Mosbach K: Molecularly imprinted monodisperse microspheres for competitive radioassay. Anal Commun 1999, 36(2):35–38. 10.1039/a809014i
Acknowledgements
The authors would like to appreciate Tehran University of Medical Sciences for funding and supporting this project. The authors also thank Mr Mirghani Seyed-Someh and Mrs Safora Arefian for their kind technical assistance through this study. The authors declare that there is no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Omidi, F., Behbahani, M., Sadeghi Abandansari, H. et al. Application of molecular imprinted polymer nanoparticles as a selective solid phase extraction for preconcentration and trace determination of 2,4-dichlorophenoxyacetic acid in the human urine and different water samples. J Environ Health Sci Engineer 12, 137 (2014). https://doi.org/10.1186/s40201-014-0137-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40201-014-0137-z