Abstract
It has become increasingly clear that both normal and cancer tissues are composed of heterogeneous populations. Genetic variation can be attributed to the downstream effects of inherited mutations, environmental factors, or inaccurately resolved errors in transcription and replication. When lesions occur in regions that confer a proliferative advantage, it can support clonal expansion, subclonal variation, and neoplastic progression. In this manner, the complex heterogeneous microenvironment of a tumour promotes the likelihood of angiogenesis and metastasis. Recent advances in next-generation sequencing and computational biology have utilized single-cell applications to build deep profiles of individual cells that are otherwise masked in bulk profiling. In addition, the development of new techniques for combining single-cell multi-omic strategies is providing a more precise understanding of factors contributing to cellular identity, function, and growth. Continuing advancements in single-cell technology and computational deconvolution of data will be critical for reconstructing patient specific intra-tumour features and developing more personalized cancer treatments.
Similar content being viewed by others
Introduction
DNA serves as the source code for specific mechanisms that regulate cellular identity, function, and growth. The genome is generally replicated with high-fidelity. However, stochastic somatic alterations can occur at an average rate of 3 mutations per cell division in normal cells [1, 2]. These genetic changes can be the effect of inherited mutations, environmental factors, or inaccurately resolved errors in transcription or replication. Mutations typically occur in non-coding regions of the genome and have no immediately apparent effect on the phenotype of the cell [2,3,4,5]. However, as mutations accumulate over time, they increase genetic variations and the likelihood of developing a neoplasm. Communities of mutations, or alterations to driver genes, can lead to increases in proliferation, a higher frequency of errors in transcription and replication, and/or the enabling of apoptotic evasion [6, 7]. Finally, recent studies indicate that metastases may also derive from early disseminated cancer cells [8]. These features are hallmarks of cancer that subsequently facilitate neoplastic progression (Fig. 1) [9].
Heterogeneity and metastasis. a Normal healthy tissues have a naturally occurring degree of somatic heterogeneity. These mutations can arise due to environmental factors and inaccurately resolved errors in transcription or replication. b As mutations stochastically arise, some will be neutral, thus having no apparent affect on the phenotype, while others may occur in ‘driver’ gene regions and have more immediately observable traits. For example, mutated DNA damage response (DDR) genes can drive tumorigenesis because they leave the cell without the necessary pathways to resolve lesions. c Driver gene mutations can confer an advantage in the founder clone and promote subsequent expansion. d Secondary mutations that occur in subclones further drive heterogeneity and can lead to metastasis. Additionally, recent research suggests that metastases may also derive from early disseminated cancer cells
To better interpret cellular heterogeneity, researchers have developed various high-throughput applications to generate a more comprehensive cellular atlas of the human body. Tang et al. [10] initially reported a single-cell RNA-seq experiment, where only one cell was sequenced in a single run. This cell was manually separated under the microscope. Since then, the technology has improved several times, each time providing a higher cell count and/or expression sensitivity in a single run. Notably, published in 2012, SMART-seq allowed for greater sensitivity and capturing of full-length transcripts, however cells had to be manually picked in that experiment limiting practical cell capture counts. The Fluidigm C1 capture method introduced microfluidic chips for more automated larger scale cell capture that could be paired with effective library preparation technologies. Starting from 2014, a number of emulsion-based protocols including that by 10× Genomics increased this number by another one to two orders of magnitude (Table 1).
Catching up with the advances in the technology, methods to investigate complex populations are only now coming to fruition with single-cell precision. For example, bulk high-throughput sequencing has been previously used to reveal that intra-tumour genetic and epigenetic heterogeneity progress through sub-clonal branched evolution rather than through linear expansion (Fig. 2) [11, 12]. However, for similar studies, single-cell tools for phylogenetic reconstruction of clonal evolution are more complicated due to lower coverage than bulk samples [13,14,15,16]. Characterizing the branched sub-clonal evolution of a neoplasm is critical for identifying key sub-population driver mutations promoting diversification, expansion, invasion, and eventually colonization to other parts of the body. In addition, the aggregated effect of tumour heterogeneity is important to resolve because resistance in one or more clonal subsets of a global tumour cellular population can impact chemotherapeutic efficacy (Fig. 2) [17]. In fact, chemotherapies have a modest overall median survival benefit of 2.1 months while costing around $100,000/year in the U.S. [18, 19]. One option to mitigate this inefficiency is to remodel patient specific intra-tumour heterogeneity computationally using single-cell genomics data and determine functional pathways at a high resolution [20,21,22]. While the circulating tumor cells provide an opportunity to directly profile the difference when comparing to the primary tumor samples retrospectively, investigating different tumor subgroups allows one to reconstruct the evolution of single tumor cells relative to each other in a more continuous fashion. To assemble a comprehensive cellular map of the body, accurate and reproducible experimental protocols, and computational analysis pipelines will be critical to extract and interpret heterogeneous information. Here, we provide a review of current single-cell genomics strategies developed for investigating cellular heterogeneity.
Clonal phylogeny in cancer and resistance. A Darwinian tree model best describes clonal evolution. a Multiregion biopsies have been used to investigate intra-tumour heterogeneity. This involves taking biopsies from different regions of the same tumour then preparing high-throughput sequencing libraries. b Phylogenetic reconstruction of clonal evolution gives a detailed understanding of heterogeneity in the tumour. A mutation occurring at the ‘trunk’ of the tree and promotes clonal expansion. Subclones arise due to subsequent mutations that diversify the population. Driver mutations can also occur later in clonal evolution and infer resistant properties that were not present in the initial driver mutation. If chemotherapy fails to knock out unique trunks, a drug-resistant population will remain and serve as the dominant feature during relapse
Single-cell partitioning
The standard workflow of single-cell investigations includes dissociating a bulk-cell sample into individual cells, isolating those cells, preparing them for the desired application, acquiring data, and analyzing data. Today, methods for separating cells still include mechanical perturbation or enzymatic digestion to separate bulk samples into single cells. However, the downstream methods for isolation, preparation, data generation, and analysis have made rapid advancements. Partitioning bulk samples and isolating individual cells can be technically challenging and necessitates optimization, often on a tissue-specific level. This initial step for investigating heterogeneity through single-cell applications can be complicated by the inefficient separation, which results in higher doublet capture rates in fluidics and droplet-based technologies. Caution should also be taken not to induce unnecessary mechanical or chemical stress on the cell during this process. Performing a clean isolation will also avoid unnecessary molecular debris that can impact the ability to assign individual unique molecular identifiers (UMIs) to single cells during the process of demultiplexing on the cellular and transcript/allele levels.
After separating and suspending the population of tumour cells from biopsy tissue, single cells can either be processed in bulk or sorted and enriched to select specific sub-populations. Most commonly, single cells can be isolated by flow-cytometry, laser capture microdissection, serial dilution, using antibody-coated magnetic beads, or microfluidic-based techniques. Droplet-based technologies such as Drop-seq, inDrop, Chromium, and ddSeq can produce tens to hundreds of thousands of uniquely barcoded cells (Table 1) [28, 35, 36]. The droplet-based approach for isolating and preparing single cells involves using bead-based surface chemistry to facilitate molecular sample preparation methods while encapsulating the cell in an emulsion or aqueous microfluidic partition (Fig. 3a). Each bead contains DNA fragments with unique barcode sequences that are incorporated with cell material during encapsulation. While encapsulated, RNA is also reverse transcribed. The emulsions are then broken prior to pooled amplification and sequencing. Integrated microfluidic circuit (IFC) chips offer an alternative approach to isolate and process cells individually by capturing them in small chambers [37]. IFC protocols have a natural quality-control step whereby doublets can be recorded by microscopy visualization before preparing downstream applications (Fig. 3b).
Single-cell isolation and preparation. a One method for isolating single-cells is by droplet microfluidics. In the first channel, individual cells are coupled with uniquely bar-coded beads that continue down the pipe until they are captured by an oil droplet. The oil droplets are then pooled in high quantities, and PCR is performed on the population. b A second approach for isolating single-cells is to pre-enrich cells by FACS then pass them through an IFC chip, which collects them into individual wells. IFC chips are available in different cell size ranges which assists in limiting the capture more than 1 cell/well. Unlike the droplet approach, PCR is performed on individual cells. This method results in lower overall cell counts than the droplet approach, however, there is a reported trade-off in sensitivity. Overall, droplet-based methods will yield higher numbers of cells per experiment, but the quality of data is more sparse, whereas, microfluidic chip methods provide a deeper cellular profile but with fewer cells. Researchers have to weigh the trade-off of single cell read depth or single-cell population breadth
Costs should be considered when deciding the satisfying number of cells to run relevant to the depth of heterogeneity in the tissue being interrogated. Isolating cell populations through a chip limits the number of cells that can be captured in a single run. It can be more expensive than droplet-based approaches (~ $3.5 versus $0.10 per cell) [38]. In addition to financial differences, these recent approaches and their derivatives each have unique technical concerns and limitations that have been reported in Svensson et al. [39]. After isolating the single-cell population of interest, the subsequent preparation steps vary widely depending on the desired application. Below, we elaborate on exemplary applications for studying single-cell heterogeneity.
Single-cell whole genome and whole exome sequencing
Fundamental technical concerns exist with generating and analyzing single-cell genetic, epigenetic, and expression data, because of the low amount of starting material. Single cells have ~ 6 pg of genomic DNA, 10–30 pg of total RNA that must be amplified before sequencing, and roughly 250–300 pg of protein that can be analyzed. A battery of computational models have been developed to address false-positives due to nonlinear amplification, false-negative allelic drop-out due to amplification bias, non-uniform coverage, and noise that arises during single-cell genome or transcript amplification (Table 2) [40]. For this reason, bioinformatics and computational biology applications are critical for analyzing NGS output files and accurately identifying genetic variation. Single-cell whole-genome and whole-exome sequencing (scWGS and scWES, respectively) provide amplified genomic DNA variant datasets that can then be used to reconstruct clonal evolution or to measure genetic heterogeneity.
Experimental methods
To date, three primary techniques are used for whole genome amplification (WGA) which include: (1) PCR-based methods such as multiple annealing and looping-based amplification cycles (MALBAC), (2) degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR); and (3) non-PCR based method using random hexamers or non-specific priming like multiple displacement amplification (MDA) [41,42,43]. Each of the three primary techniques becomes unreliable when there is less than 1.5 pg of genomic material. The smaller starting volumes are susceptible to environmental contamination and require optimal sterile working conditions to avoid the creation of false positives [44]. Each method used for single-cell genomic DNA amplification has biases to consider and can be affected by poor coverage or uneven sequencing depth, which will result in noisy and inaccurate read counts.
MALBAC tends to have GC bias, but reports find that MALBAC libraries are more reproducible than those generated with MDA [45]. Some analytic tools, such as the Ginkgo web platform (http://qb.cshl.edu/ginkgo), take measures to reduce or eliminate these amplification biases. According to the authors of Ginkgo, of the three genetic amplification techniques used, MDA has non-uniform coverage and worse GC bias than either DOP-PCR or MALBAC. Reports suggest that despite moderate physical coverage DOP-PCR is the most reliable method [42, 46]. While this Review was being prepared, an approach that uses linear amplification via transposon insertion (LIANTI) was reported that may prove to have less amplification bias and high (97%) genome coverage [47].
Exemplary applications
With new isolation, amplification, and computational modelling techniques becoming available there has been a general progression toward increased populations of cells or higher quality coverage in rare populations. In 2012, scWGA by MDA of 58 cells was used to demonstrate mutational contributions of SESN2 and NTRK1 in neoplasm progression. In this study, more than 90% of the single-cell genomic data mapped back to the reference genome and they had an 11% allele dropout rate [48]. In another early example, Xu et al. performed scWES-seq of 25 single cells by MDA amplification which was used to reconstruct clonal mutations occurring within PBRM1 and VHL in kidney cancer. Here, the MDA approach yielded a false positive rate of 2.67 × 10−5 and an allele dropout rate of 16.43% [49]. A more recent study by Gao et al. used DOP-PCR to amplify genomic content from 1000 cells and reported that the majority of subclonal CNV occurs shortly after the onset of the primary driver mutation in breast cancer. While the false discovery rate was not mentioned in this article, their data suggests that 20–40 single cells were required for detecting subpopulations with 95% power [50]. It is clear that the number of cells being reported in a single study has changed rapidly. However, while the available number of cells per analysis has been used to reach up to nearly 1500 cells (using MDA), the average number of reads per cell decreases as the number of cells increase. Thus, much like scRNA-seq, there is a tradeoff between the individual cell quality and the total number of cells analyzed within a batch [39]. The shortcoming of large batches emphasizes the need for computational tools that can correct such bias (Table 2).
Single-cell RNA sequencing
The most broadly developed method for single-cell analysis is single-cell RNA sequencing (scRNA-seq). High-throughput RNA-seq of bulk samples from scores of patients have provided novel insights into many cancer types [51,52,53]. However, deploying RNA-seq analysis at single-cell resolution can provide an even deeper level of understanding the heterogeneous composition of tumour samples by identifying constituents otherwise masked in bulk RNA-seq [54, 55]. The first scRNA-seq was performed in 2009 using single mouse embryo blastomeres [10]. Since then, there has been an increasing surge of sc-RNA-seq publications. Compared to bulk level RNA-seq, scRNA-seq has at least two advantages. First, a more accurate and sensitive presentation of the cell-to-cell variability can be discerned. Second, these data can be reorganized into pseudo-temporal arrangements that can more accurately reconstruct clonal evolution. Beyond experimental advantages, monitoring the dynamics of sub-clonal populations across the course of treatment also has the potential to inform and allow more precise adjustment of therapies.
Experimental and computational methods
There are several platforms available for isolating and preparing RNA from single cells. One of the greatest technical concerns is in amplifying these low concentrations without introducing significant bias. Data generated by droplet-based approaches permit 10,000 s of cells to be counted, while other platforms that use chip-based systems process only 100 s of cells at a time but are more sensitive to calling the number of genes per cell [39]. After successful isolation of single cells, there are a wide number of molecular approaches to creating scRNA-seq libraries such as SMART-seq, SUPeR-seq, BAT-seq, CEL-seq, and STRT-seq amongst others. The SMART-seq approach can generate full-length cDNA, whereas approaches like STRT-seq (targeting 5′ end of mRNA) and CEL-seq (targeting 3′ end of mRNA) can be used for multiplexing samples [23, 25, 27]. SMART-seq employs a special reverse-transcriptase that anchors both ends of cDNA with distinct nucleotides. The absence of one of these ends eliminates incompletely reverse-transcribed RNA molecules after a subsequent cloning step. It is important to note that sensitivity and accuracy become concerns when the experiments scale up to larger numbers. Here, sensitivity is defined as the smallest quantity of input RNA molecules required for a gene to be confidently called. Accuracy is defined here as the closeness between the estimated and the actual abundance levels of input molecules. Whereas studies have shown that the droplet-based microfluidic approaches permit greater numbers of cells to be counted, chip-based systems appear to be more sensitive [39].
Computationally, many tools have been tailored to take advantage of the high-resolution of scRNA-seq data and deconvolute noise [56]. For example, Monocle2 is an unsupervised algorithm designed to analyze the heterogeneity among cells and reconstruct the micro-evolution timeline from scRNA-seq data [57]. Other tools such as scLVM [58], PseudoGP [59], and SPADE [60] have provided various solutions to analyze heterogeneity with scRNA-seq data computationally. With the scRNA-seq analysis toolbox expanding rapidly, graphical user interface (GUI) pipelines such as Granatum (http://garmiregroup.org/granatum/app) have recently been developed to ensure that accessing the latest development in computational methods is amenable for clinical and non-informatics researchers [61]. In addition, with datasets accumulating at an astonishing speed, there have been efforts like the RIKEN Single-Cell Project (http://singlecell.riken.jp/en/) to consolidate, index, and organize publically available datasets [62].
Exemplary applications
scRNA-seq has been used broadly to provide data on genetic expression and has now been widely applied to a variety of cancer types. Since there are many more techniques developed and reported for scRNA-seq, we will only highlight a few applications as examples of how scRNA-seq is impacting the discussion on heterogeneity. In one example, the SMART-seq protocol was used to profile full-length mRNA from 430 primary glioblastomas to reveal an intratumor spectrum of differentiation states [63]. The SMART-seq protocol was later improved to increase mRNA yield, coverage, sensitivity, accuracy and reintroduced as Smart-seq 2 [27]. Smart-seq 2 is now a widely used approach in scRNA-seq. In one case, it was used to profile 4347 cells from six oligodendrogliomas and revealed subgroups of undifferentiated cells with a stem-cell-like expression that may be the source of oligodendrogliomas [54]. These data highlight that one benefit of performing expression analysis at single-cell resolution is it can reveal subpopulations otherwise masked in bulk data. In addition, enhancing sensitivity for clonal-level therapies alongside offers the potential for discovering novel, previously undetectable biomarkers on an individual level.
Currently, one of the most interesting shifts in research to recognize is that scRNA-seq is progressing to include a broader range of samples in addition to a deeper pool from a single source. Thus, as the cost of performing scRNA-seq continues to drop, it will facilitate simultaneous inter- and intra-tumour investigations. For example, a recent report using 9879 cells from 10 IDH-A tumours and 4347 cells from six IDH-O tumours were compared with 165 TCGA bulk samples to identify a common progenitor for IDH mutant gliomas [64]. This report is representative of the growing trend to combine available bulk data with single-cell data cohorts for broader and deeper data mining potential. In addition to profiling tissue samples, scRNA-seq is also used to investigate circulating tumour cells (CTC), which is particularly valuable for prospective monitoring [65]. The SMART-seq method was initially reported in an investigation on CTCs and was proposed as a method for identifying candidate cancer biomarkers [24]. Isolating and analyzing expression in CTCs alongside monitoring circulating cell-free DNA (ccfDNA) burden has high direct translational potential for identifying personalized biomarker panels to guide treatment in real-time.
Single-cell chromosome conformation capture
Developments in studying single-cell genomic architecture have become increasingly deployed to understand the relationship between topology and phenotype. Topology is interesting because single nucleotide variations (SNV), point mutations, and insertions or deletions (indels) can indirectly impact the expression of a distant gene by rearranging the genetic architecture [66]. Since the expression of some genes is affected by long-distance interactors, another level of heterogeneity to consider is the arrangement and proximity of chromosome territories within the nucleus [67]. Genomic architecture is described as being organized in A/B compartments, topologically associated domains (TADs), and loops. Perturbation at any level of these structures can have a significant role in determining domain accessibility which can either improve or inhibit activity in that region. These physical genomic folding structures have been revealed using long-range genomic interaction maps derived from high-throughput sequencing data [68]. When integrated with other techniques such as scWES or single-cell RNA-seq, Hi-C provides an informative tool for identifying the relationship between the 3D architecture of the genome and gene expression [69].
Experimental methods
To analyze single-cell nuclear DNA structure, high-throughput sequencing is coupled with a high-resolution chromatin conformation capture (3C) assay (sciHi-C) [70]. The chromatin architecture of single cells is reconstructed by generating short and long-range interaction maps. Briefly, interaction is inferred by fixing the DNA, followed by enzymatic restriction digestion, adaptor ligation, and proximity ligation. This sequence of steps allows interacting loops and TADs to be ligated together and will thereby yield a higher number of reads due to more frequent interaction [71]. Pipelines for analyzing multiplexed scHi-C data have recently become open-sourced [71]. On a more local scale, single-cell ATAC-seq (scATAC-seq) is used to profile open (transcription-permissive) chromatin. During scATAC-seq, isolated nuclei are processed by Tn5 tagmentation, which inserts adapters into nucleosome-free regions [72, 73].
Exemplary applications
scHi-C methods give detailed information about the state of the chromatin accessibility and long-range interactions. These methods are currently being adopted to define how architecture evolves throughout the cell-cycle at single-cell resolution [74]. A recent publication used scHi-C to reveal the architecture during pronuclear formation of G1 zygotes. In the zygote, maternal and paternal pronuclei have different levels of organization with maternal DNA lacking A/B compartments. This suggests that organization of paternal compartmentalization is likely inherited from the sperm [75]. The organization of loops, TADs, and compartments is significant because it infers which regions are more active and perhaps more prone to mutations [7]. On a more localized level, a separate study used a modified sciATAC-SEQ approach called SCI-seq was demonstrated on 16,000 single cells from different cancer types. SCI-seq uses a lithium-assisted nucleosome depletions strategy to remove histones followed by cross-linking than by the scATAC-seq protocol [76]. Together, these techniques provide information on the organization of DNA in the nucleus.
Single-cell epigenetics
Epigenetic diversity involves heritable changes that affect genomic expression but that do not affect the DNA information. This includes direct modification of nucleic acids (i.e. 5mC, 6mA, m6A, and pseudouridine), and post-translational modification of histones (e.g. methylation, and acetylation) [77, 78]. DNA methylation adds another layer of complexity to our understanding of how heterogeneity affects cellular identity and function. Hypermethylation of DNA is associated with transcriptional repression, while the reverse is true for hypomethylation. Single-cell epigenetics studies have advanced more rapidly than those that deal with proteins. Elucidating epigenetic heterogeneity at the single-cell level add a deeper understanding of how methylation patterns are maintained on a clonal level across cell populations and across individuals. However, it should be noted that the most significant roadblock to generating single-cell resolution methylation data is that current techniques are harsh and result in massive loss of DNA template. Also, these techniques often include amplification methods that remove the ability to detect epigenetic modifications.
Experimental methods and applications
Amongst the experimental approaches inferring cytosine modification, single-cell bisulfite sequencing (scBS) is the most widely used technique. scBS-seq preferentially de-aminates unmethylated cytosine thereby converting unmethylated cytosines to thymines. However, during this bisulfite treatment step, nicks and fragmentation in the DNA occur that reduce the quality and quantity of the input. This is followed by primary and secondary adapter ligation and PCR [79,80,81]. Single-cell reduced representation bisulfite sequencing (scRRBS) has lower coverage of total CpG sites but higher coverage of CpG islands [82]. Aligned reads generated from this technique require special tools such as Bismark for read mapping and methylation calling [83]. A third technique known as single-cell whole-genome bisulfite sequencing (scWGBS-seq) was developed that does not include the pre-amplification step that takes place in scBS-seq but this approach has lower coverage complexity [84]. Comparatively, the scRRBS method only covers 1% CpG sites across the genome, in contrast to 48.4% of CpG sites by scWGBS. Single-cell methylase assisted bisulfite sequencing (scMAB-seq) and CpG island methylation sequencing for single-cell (scCGI-seq) have also recently been proposed [85, 86]. Future comparative studies of these various methods will assist in determining the differences in mapping and cost efficiency.
Cytosine methylation studies on cancer samples at the single-cell level currently lag behind other -omics approaches. The technical difficulties of bisulfite treatment yield poor coverage, thus these methods might not be able to unveil untargeted features of different cell populations. For this reason, Farlik et al. inferred cell line drug response in developing the scWGBS approach [84]. Finally, recent single-cell studies proposed different analytical tools to correct the input features, such as the use of a deep neural network to reconstruct noisy and missing CpG data [87]. Using computational methods to fill in sparse data will not only rescue poorly resolved data but may also be applied to identify biomarkers, project clonal evolution, or rank potential drug responses.
Single-cell proteomics
There are many layers to deciphering cell-to-cell heterogeneity. Since there is not a direct 1:1 turnover of mRNA occurrence to protein translation, adopting single-cell proteomic studies provides information on the final layer of inter- and intra-tumour heterogeneity. Unlike genetic or expression analysis, proteomic investigations at the single-cell level lack a way to amplify the starting material. Therefore, single-cell proteomic studies have the technical challenge of developing more sensitive methods for detection.
Experimental and computational methods
Quantifying proteins at single-cell resolution is a developing technology complicated by the transient nature of functional proteins. Much like other -omics sections covered earlier, investigating the proteome at single-cell resolution requires accounting for low input levels [88]. Single-cell time-of-flight mass cytometry (CyTOF) is one method used to address this issue. CyTOF targets epitopes of interest utilizing antibodies coupled with transient metal element isotopes. Single-cell droplets are introduced to inductively coupled argon plasma where the cell is vaporized, and the atomic constituents are ionized before time-of-flight sampling [89]. Since CyTOF is limited to around 50 parameters, this approach yields a much lower throughput than scRNA-seq. However, it is more affordable than scRNA-seq and can help to determine if an enrichment (e.g. FACS) step is necessary before transcriptome analysis. Another mass spectrometry approach recently proposed is a method known as single-cell proteomics by mass spectrometry (SCoPE-MS). The SCoPE-MS workflow was designed to isolate protein from single-cells and prepare each cell for MS. SCoPE-MS attempts to resolve the issue of protein loss during transfer and low starting material by manually separating and lysing cells. This method includes tandem mass tags for reporter ion relative abundance quantification [90]. Alternatively, non-MS approaches for single-cell proteomic studies can utilize chip-based isolation. In the single-cell barcoded chip (SCBC) method, cells are isolated into wells and probed with antibody arrays which are analyzed by a microarray scanner [91]. Antibody arrays are also utilized in single-cell western blotting (scWB) for which isolated cells are lysed, and SDS-PAGE is applied to each well. Relative to the MS approaches, scWB is limited to probing the sample with a smaller panel of antibodies.
There are a variety of statistical tools available for inferring subpopulations and subpopulation specific markers from single-cell protein data including SPADE, Phenograph, and Wishbone [92]. For example, histoCAT is a new powerful integrative method used to integrate single-cell CyTOF measurement with image-based spatial information to detect spatial and phenotype interaction at the cellular level [93]. Also, new studies propose different methods to construct dynamic protein signalling networks using single-cell protein measurement [94, 95]. In particular, one new approach created a dynamic regulation network from CyTOF measurements to model the drug perturbation of the epithelial-to-mesenchymal transition [96]. This type of approach can then facilitate the discovery of critical events correlated with a cell state transition.
Exemplary applications
One example of the SCBC method for single-cell chip-based proteomic investigation first heavily dilutes a FACS enriched population before loading them into microchambers. Wei et al. used a phosphoproteomic antibody array to profile mTORki resistant glioblastomas [97]. This SCBC study, cells were lysed after isolation, and the protein contents were captured using custom antibody barcodes. Chip-based methods can assay a relatively larger number of proteins (n > 40) than other techniques currently available, but still fewer the MS approaches [88]. For this reason, the continuing development of single-cell mass spectrometry strategies such as CyTOF will be the technology that unlocks single-cell proteomic scalability. As a recent example, a panel of more than 30 antibodies was used in CyTOF to analyze tumour cells, adjacent normal tissue, and blood from 28 patients with lung adenocarcinoma [98]. The CyTOF data reported here revealed that tumour-infiltrating myeloid cells likely shape the composition of anti-tumour T-cells. Thus, the ability to profile large population of single-cell surface markers has powerful implication in immunology.
Single-cell multi-omics
Single-cell multi-omic strategies capture the most accurate state of factors that contribute to the cellular phenotype. Ultimately, the integration of several layers of data will be necessary for deconvoluting the relationship between expression, function, and identity. This is because bulk level analysis can only describe the general trends in a population that can mask cellular subtypes [99]. Multi-omic studies are complicated by the technical requirement of separating and preserving different molecular layers from the same cell. Bioinformatics and computational biology provide critical support for reconstructing features that may become noisy as a result of sample loss during multi-omic sample preparation.
Experimental methods
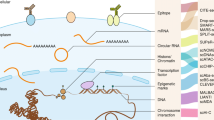
The most cutting-edge research calls for investigators to combine all of the techniques discussed to reconstruct multi–omic single-cell profiles (Fig. 4). Achieving this level of resolution will provide the most comprehensive profile of cell-to-cell diversity in normal and tumour tissue and inform researchers on the impact of single-cell genetic and epigenetic heterogeneity. Bock et al. proposed that molecules collected from the same cell can be assayed by one of several approaches depending on the desired downstream application [100]. For example, two methods described involve either separating the molecular layers (e.g. DNA and RNA) into their equivalents or splitting the sample itself into different fractions and proceeding with the desired isolation within the given fraction. Alternatively, multi-omic methods can be combined into a single workflow [100]. Taken together, the cluster of data generated by multi-omic approaches can infer the underlying triggers of cellular identity and function. As these techniques in cell isolation and amplification continue to improve, multiple layers of heterogeneity reconstruction can be used to identify neoplastic predisposition markers and provide a refined map for precise drug treatment regimen.
Single-cell multi-omics analysis workflow. a Multi-omic technologies can produce reads from the transcriptome (RNA-seq), the genome (exome sequencing), and/or the methylome, from the same cells. b Read alignments, quality control (QC), and specific processing steps create “feature expression” matrices, where cells are represented as vectors and genomic features (e.g. gene expression, methylation) represented as columns. c The different omic matrices can then be analyzed independently, for detecting cell subpopulations and ranking the genomic features etc. d Finally, multi-omics integration can be performed to identify coherent features from different omics that separate different subpopulations
Exemplary applications
Since multi-omic strategies are at the frontier of single-cell research, the body of literature remains relatively nascent. The majority of current available single-cell multi-omics technologies are focused on the link between epigenetic and transcriptional variations. Macaulay et al. established scG&T-seq to simultaneously measure genetic variation and gene expression via separation of gDNA from polyA RNA using a biotinylated oligo-dT primer [101]. scG&T-seq equips Smart-seq 2 for whole transcriptome analysis and offers various methods for DNA amplification depending on the downstream application (MDA, PicoPlex etc.). In contrast to conventional scRNASeq sequencing methods, scG&T-seq utilizes ERCC-spike-ins to assess the number of genes expressed and transcript coverage lengths. Angermueller et al. developed another method called scM&T-seq to evaluate the relationship between methylation and transcription variations in heterogeneous cell populations through scRNASeq and scBS-seq techniques [102]. scM&T-seq was applied in discriminating 61 mouse serum ESCs (embryonic stem cells) and 16 ESCs grown in 2i media. The results showed that the connection strength between methylome and transcriptome varies from cell to cell. Another similar method called scMT-seq applied Smart-seq 2 and scRRBS for single-cell transcriptome sequencing and methylome sequencing, respectively [103]. Compared to scM&T-seq, scMT-seq provides similar CpG islands overlap in a more cost-effective way. This study helped to reveal the relationship of gene expression and DNA methylation in gene body and promoter regions in neuron single cells.
Regarding simultaneous measurement of gene expression with another omics data generated from the same cell, DR-Seq is an example of pioneering work on single-cell DNA and RNA parallel sequencing [104]. Without manually separating the nucleus and cytosolic mRNA, DR-Seq applies a quasi-linear amplification method with predefined adaptors to quantify gDNA and mRNA. Additionally, the comparison of DR-Seq and CEL-Seq showed that the additional steps for amplification of gDNA would not affect the mRNA results. However, this single-pot strategy requires in silico masking of the coding sequences (exonic region) of the genome to determine copy number variation, which leads to incomplete transcripts from the cell. Another recently published work by Stoeckius et al. developed CITE-seq to integrate cellular protein markers and transcriptome in single cells through oligonucleotide-labeled antibodies [105]. CITE-Seq not only enabled to differentiate cellular subgroups based on surface protein expression, but also achieved a consistent output of protein detection with currently standardized flow cytometry. Compared to scRNA-seq alone, CITE-Seq demonstrates both the highly consistent protein and RNA profiles with literature and also an enhancement of characterization of cell phenotypes based on immune cell experiments.
From the analytical perspective, certain additional layers can be reconstructed given one layer is provided. For example, SNVs can be directly extracted from RNA-seq reads and then correlated to gene expression [106]. In addition, by using the input fastq files it is now possible to highlight predictive DNA motifs linked to methylation profiles [87]. However, most of the high-throughput scRNA-seq pipelines are designed specifically for mRNA expression counting. Since the library products are quite short, there is fairly limited capability to do this without modifying more commercial protocols.
Three-omics single-cell assays also became possible. Recently, Hou et al. established scTrio-seq to simultaneously sequence and analyze single cell’s genomic copy number variations, DNA methylation and transcriptomic gene expression together [107]. scTrio-seq demonstrated its ability to efficiently measure DNA methylome, transcriptome and genome copy number compared to scRRBS, bulk cell RNA-seq and bulk cell RRBS and bulk cell WGBS. The integration of triple omics information via scTrio-seq on 25 HCC cancer cells identifies two heterogeneous subpopulations with different malignancy and metastasis potential.
Conclusions
High-throughput sequencing techniques provide clinicians with a more comprehensive understanding of the genetic and epigenetic heterogeneity in normal and cancer cells. Moreover, future personalized treatments might integrate as a routine single-cell strategy to unveil intra-tumour heterogeneity and thus provide a more accurate therapy regimen. Multi-omics approaches that detail inter- and intra-tumour heterogeneity within individual patients will continue to evolve and provide critical insight to informing more accurate treatment regimens based on prognosticated drug response. In addition, these emerging molecular techniques when combined with computational analysis tools will advance research in other areas such as developmental biology, biotechnology, pathology and more. As larger amounts of single-cell data become publicly available, there will be increased opportunities to identify subclonal-specific biomarkers at a personalized level. User-friendly data portals for single-cell analysis, such as Granatum, will become increasingly integral in the bench-to-bedside transition [61]. The comprehensive annotation and analysis of single-cell datasets will be the foundation of understanding how cell-to-cell variability in normal and cancer cells influence cellular identity and function in the human body.
Abbreviations
- 3C:
-
chromosome conformation capture
- BAT-seq:
-
barcoded 3′-end specific sequencing
- BWA:
-
Burrows-Wheeler Aligner
- ccfDNA:
-
circulating cell-free DNA
- CD:
-
cluster of differentiation
- CEL-Seq:
-
cell expression by linear amplification and sequencing
- CIViC:
-
clinical interpretation of variants in cancer
- CNV:
-
copy number variation
- CNVkit:
-
copy number variation kit
- CTC:
-
circulating tumor cells
- ddSeq:
-
droplet digital sequencing
- DOP-PCR:
-
degenerate oligonucleotide primed PCR
- ECdetect:
-
extrachromosomal DNA
- EwS:
-
Ewing sarcoma
- GATK:
-
genome analysis tool kit
- GUI:
-
graphical user interface
- IDH:
-
isocitrate dehydrogenase
- iEVORA:
-
epigenetic variable outliers for cancer risk prediction analysis
- IFC:
-
integrated microfluidic circuit
- InferCNV:
-
infer copy number variation
- INTEGRATE-Neo:
-
integrate neoantigens
- LIANTI:
-
linear amplification via transposon insertion
- MALBAC:
-
multiple annealing and looping based amplification cycles
- MARS-seq:
-
massively parallel single-cell RNA-sequencing
- MDA:
-
multiple displacement amplification
- MS:
-
mass spectrometry
- mTORki:
-
mTOR kinase inhibitor
- MutSigCV 2.0:
-
mutation significance copy variation
- NTRK1:
-
neurotrophic receptor tyrosine kinase 1
- OncoNEM:
-
oncogenetic nested effects model
- scATAC-seq:
-
single-cell assay for transposase accessible chromatin sequencing
- scBS:
-
single-cell bisulfite sequencing
- scCGI-seq:
-
CpG island methylation sequencing for single-cell
- sciHi-C:
-
high-resolution chromatin conformation capture (3C) assay
- SCITE:
-
single-cell inference of tumor evolution
- scLVM:
-
single-cell latent variable models
- scMAB-seq:
-
single-cell methylase assisted bisulfite sequencing
- scMT-seq:
-
single-cell methylome and transcriptome sequencing
- SCoPE-MS:
-
single-cell ProtEomics by mass spectrometry
- scRNA-seq:
-
single-cell RNA sequencing
- scRNASeqDB:
-
single-cell RNA sequencing database
- scRRBS:
-
single-cell reduced representation bisulfite sequencing
- scWES:
-
single-cell whole exome sequencing
- scWGBS-seq:
-
single-cell whole genome bisulfite sequencing
- scWGS:
-
single-cell whole genome sequencing
- SESN2:
-
sestrin 2
- SMART-seq:
-
switching mechanism at the 5′ end of the RNA transcript sequencing
- SNP:
-
single-nucleotide polymorphism
- SPADE:
-
spanning-tree progression analysis of density-normalized events
- SPLiT-Seq:
-
split pool ligation-based transcriptome sequencing
- SSrGE:
-
sparse SNV inference to reflect gene expression
- STRT-Seq:
-
single-cell tagged reverse transcription
- SUPeR-seq:
-
single-cell universal poly(A)-independent RNA sequencing
- TAD:
-
topologically associated domains
- TCGA:
-
The Cancer Genome Atlas
- UMI-seq:
-
unique molecular identifier sequencing
- WGA:
-
whole genome amplification
References
Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R et al (2005) A quantitative measurement of the human somatic mutation rate. Cancer Res 65(18):8111–8117
Tomasetti C, Vogelstein B, Parmigiani G (2013) Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci 110(6):1999–2004
Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N et al (2016) Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538(7624):260–264
Tomasetti C, Li L, Vogelstein B (2017) Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355(6331):1330
Tak YG, Farnham PJ (2015) Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8(1):57
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW (2013) Cancer genome landscapes. Science 339(6127):1546–1558
Fagny M, Paulson JN, Kuijjer ML, Sonawane AR, Chen C-Y, Lopes-Ramos CM et al (2016) A network-based approach to eQTL interpretation and SNP functional characterization. bioRxiv. https://www.biorxiv.org/content/early/2016/11/09/086587
Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R et al (2016) Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540:588
Robinson DR, Wu Y-M, Lonigro RJ, Vats P, Cobain E, Everett J et al (2017) Integrative clinical genomics of metastatic cancer. Nature 548(7667):297–303
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N et al (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6(5):377–382
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366:883–892
Cheow LF, Courtois ET, Tan Y, Viswanathan R, Xing Q, Tan RZ et al (2016) Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat Methods 13(10):833–836
Jahn K, Kuipers J, Beerenwinkel N (2016) Tree inference for single-cell data. Genome Biol 17(1):86
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J et al (2011) Tumour evolution inferred by single-cell sequencing. Nature 472(7341):90–94
Smith MA, Nielsen CB, Chan FC, McPherson A, Roth A, Farahani H et al (2017) E-scape: interactive visualization of single-cell phylogenetics and cancer evolution. Nat Methods 14(6):549–550
Salehi S, Steif A, Roth A, Aparicio S, Bouchard-Côté A, Shah SP (2017) ddClone: joint statistical inference of clonal populations from single cell and bulk tumour sequencing data. Genome Biol 18(1):44
Schmidt F, Efferth T (2016) Tumor heterogeneity, single-cell sequencing, and drug resistance. Pharmaceuticals 9(2):33
Prasad V, De Jesus K, Mailankody S (2017) The high price of anticancer drugs: origins, implications, barriers, solutions. Nat Rev Clin Oncol 14(6):381–390 (advance online publication)
Fojo T, Mailankody S, Lo A (2014) Unintended consequences of expensive cancer therapeutics—the pursuit of marginal indications and a me-too mentality that stifles innovation and creativity: the John Conley lecture. JAMA Otolaryngol Head Neck Surg 140(12):1225–1236
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I et al (2014) Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46(3):225–233
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS et al (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481(7382):506–510
Schwartz R, Schaffer AA (2017) The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet 18(4):213–229
Islam S, Kjällquist U, Moliner A, Zajac P, Fan J-B, Lönnerberg P et al (2011) Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21(7):1160–1167
Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR (2012) Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30:777–782
Hashimshony T, Wagner F, Sher N, Yanai I (2012) CEL-Seq: single-cell RNA-seq by multiplexed linear amplification. Cell Reports 2(3):666–673
Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M et al (2014) Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11(2):163–166
Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R (2013) Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10(11):1096–1098
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R et al (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8:14049
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M et al (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161(5):1202–1214
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V et al (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161(5):1187–1201
Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM et al (2016) Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34(3):303–311
Taylor K, Watson L, Frenz L, Greiner D, Lebofsky R, Do D et al (2017) A scalable high-throughput method for RNA-seq analysis of thousands of single cells. https://jp.illumina.com/content/dam/illumina-marketing/documents/products/flyers/ddseq-single-cell-poster-handout-single-cell-poster-handout-web.pdf. Accessed 18 Dec 2017
Rosenberg AB, Roco C, Muscat RA, Kuchina A, Mukherjee S, Chen W et al (2017) Scaling single cell transcriptomics through split pool barcoding. bioRxiv. https://www.biorxiv.org/content/early/2017/02/02/105163
Gierahn TM, Wadsworth MH II, Hughes TK, Bryson BD, Butler A, Satija R et al (2017) Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 14(4):395–398
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M et al (2015) Highly parallel genome-wide expression profiling of individual CELLS using nanoliter droplets. Cell 161(5):1202–1214
Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM et al (2017) Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 12(1):44–73
Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR (2000) Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288(5463):113
Papalexi E, Satija R (2017) Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. https://doi.org/10.1038/nri.2017.76 (advance online publication)
Svensson V, Natarajan KN, Ly L-H, Miragaia RJ, Labalette C, Macaulay IC et al (2017) Power analysis of single-cell RNA-sequencing experiments. Nat Methods 14(4):381–387
Huang L, Ma F, Chapman A, Lu S, Xie XS (2015) Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet 16:79–102
Chapman AR, He Z, Lu S, Yong J, Tan L, Tang F et al (2015) Single cell transcriptome amplification with MALBAC. PLoS ONE 10(3):e0120889
Garvin T, Aboukhalil R, Kendall J, Baslan T, Atwal GS, Hicks J et al (2015) Interactive analysis and assessment of single-cell copy-number variations. Nat Methods 12(11):1058–1060
Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13(3):718–725
de Bourcy CFA, De Vlaminck I, Kanbar JN, Wang J, Gawad C, Quake SR (2014) A quantitative comparison of single-cell whole genome amplification methods. PLoS ONE 9(8):e105585
Ning L, Li Z, Wang G, Hu W, Hou Q, Tong Y et al (2015) Quantitative assessment of single-cell whole genome amplification methods for detecting copy number variation using hippocampal neurons. Sci Rep 5:11415
Wang Y, Navin NE (2015) Advances and applications of single cell sequencing technologies. Mol Cell 58(4):598–609
Chen C, Xing D, Tan L, Li H, Zhou G, Huang L et al (2017) Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI). Science 356(6334):189–194
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X (2012) Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148:873–885
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L (2012) Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148:373–385
Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y et al (2016) Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 48(10):1119–1130
The Cancer Genome Atlas Research Network (2017) Integrated genomic and molecular characterization of cervical cancer. Nature 543(7645):378–384
The Cancer Genome Atlas Research Network (2017) Integrated genomic characterization of oesophageal carcinoma. Nature 541(7636):169–175
Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J et al (2017) Integrative clinical genomics of metastatic cancer. Nature 548(7667):297–303
Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K et al (2016) Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539(7628):309–313
Yang J, Tanaka Y, Seay M, Li Z, Jin J, Garmire LX et al (2017) Single cell transcriptomics reveals unanticipated features of early hematopoietic precursors. Nucleic Acids Res 45(3):1281–1296
Zhu X, Ching T, Pan X, Weissman SM, Garmire L (2017) Detecting heterogeneity in single-cell RNA-Seq data by non-negative matrix factorization. PeerJ 5:e2888
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M et al (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32(4):381–386
Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ et al (2015) Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechol 33(2):155–160
Campbell KR, Yau C (2016) Order under uncertainty: robust differential expression analysis using probabilistic models for pseudotime inference. PLoS Comput Biol 12(11):e1005212
Anchang B, Hart TD, Bendall SC, Qiu P, Bjornson Z, Linderman M et al (2016) Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protoc 11(7):1264–1279
Zhu X, Wolfgruber TK, Tasato A, Arisdakessian C, Garmire DG, Garmire LX (2017) Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists. Genome Med 9(1):108
Cao Y, Zhu J, Han G, Jia P, Zhao Z (2017) scRNASeqDB: a database for gene expression profiling in human single cell by RNA-seq. bioRxiv. https://www.biorxiv.org/content/early/2017/01/31/104810
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344(6190):1396–1401
Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG et al (2017) Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355(6332):eaai8478. https://doi.org/10.1126/science.aai8478
Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y et al (2016) HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537(7618):102–106
Lupianez DG, Spielmann M, Mundlos S (2016) Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet 32(4):225–237
Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2(4):292–301
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A et al (2009) Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 326(5950):289–293
Seaman L, Chen H, Brown M, Wangsa D, Patterson G, Camps J et al (2017) Nucleome analysis reveals structure-function relationships for colon cancer. Mol Can Res 15(7):821–830
Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W et al (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502(7469):59–64
Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM et al (2017) Massively multiplex single-cell Hi-C. Nat Methods 14(3):263–266
Cusanovich DA, Daza R, Adey A, Pliner H, Christiansen L, Gunderson KL et al (2015) Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348(6237):910–914
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10(12):1213–1218
Nagano T, Lubling Y, Várnai C, Dudley C, Leung W, Baran Y et al (2017) Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547(7661):61-67
Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N et al (2017) Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544(7648):110–114
Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH et al (2017) Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14(3):302–308
Chen K, Zhao BS, He C (2016) Nucleic acid modifications in regulation of gene expression. Cell Chem Biol 23(1):74–85
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21(3):381–395
Miura F, Ito T (2015) Highly sensitive targeted methylome sequencing by post-bisulfite adaptor tagging. DNA Res 22(1):13–18
Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J (2014) Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 11:817–820
Clark SJ, Smallwood SA, Lee HJ, Krueger F, Reik W, Kelsey G (2017) Genome-wide base-resolution mapping of DNA methylation in single cells using single-cell bisulfite sequencing (scBS-seq). Nat Protoc 12(3):534–547
Guo H, Zhu P, Guo F, Li X, Wu X, Fan X (2015) Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc 10:645–659
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics 27(11):1571–1572
Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schonegger A, Klughammer J et al (2015) Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep 10(8):1386–1397
Wu X, Inoue A, Suzuki T, Zhang Y (2017) Simultaneous mapping of active DNA demethylation and sister chromatid exchange in single cells. Genes Dev 31(5):511–523
Han L, Wu HJ, Zhu H, Kim KY, Marjani SL, Riester M et al (2017) Bisulfite-independent analysis of CpG island methylation enables genome-scale stratification of single cells. Nucleic Acids Res 5:e77
Angermueller C, Lee HJ, Reik W, Stegle O (2017) DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol 18(1):67
Heath JR, Ribas A, Mischel PS (2016) Single cell analytic tools for drug discovery and development. Nat Rev Drug Discov 15(3):204–216
Bendall SC, Simonds EF, Qiu P, el Amir AD, Krutzik PO, Finck R et al (2011) Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332(6030):687–696
Budnik B, Levy E, Slavov N (2017) Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. bioRxiv. https://www.biorxiv.org/content/early/2017/03/15/102681
Shi Q, Qin L, Wei W, Geng F, Fan R, Shin YS et al (2012) Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Natl Acad Sci USA 109(2):419–424
Su Y, Shi Q, Wei W (2017) Single cell proteomics in biomedicine: high-dimensional data acquisition, visualization, and analysis. Proteomics. https://doi.org/10.1002/pmic.201600267
Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D et al (2017) histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods 14(9):873-876
Marr C, Zhou JX, Huang S (2016) Single-cell gene expression profiling and cell state dynamics: collecting data, correlating data points and connecting the dots. Curr Opin Biotechnol 39:207–214
Mukherjee S, Stewart D, Stewart W, Lanier LL, Das J (2017) Connecting the dots across time: reconstruction of single-cell signalling trajectories using time-stamped data. R Soc Open Sci 4(8):170811
Krishnaswamy S, Zivanovic N, Sharma R, Pe'er D, Bodenmiller B (2017) Learning edge rewiring in EMT from single cell data. bioRxiv. https://www.biorxiv.org/content/early/2017/06/25/155028
Wei W, Shin Young S, Xue M, Matsutani T, Masui K, Yang H et al (2016) Single-cell phosphoproteomics resolves adaptive signaling dynamics and informs targeted combination therapy in glioblastoma. Cancer Cell 29(4):563–573
Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C et al (2017) Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169(4):750.e17–765.e17
Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL et al (2017) Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 49(5):708–718 (advance online publication)
Bock C, Farlik M, Sheffield NC (2017) Multi-omics of single cells: strategies and applications. Trends Biotechnol 34(8):605–608
Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ et al (2015) G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods 12(6):519–522
Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX et al (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13(3):229–232
Hu Y, Huang K, An Q, Du G, Hu G, Xue J et al (2016) Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol 17(1):88
Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A (2015) Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 33(3):285–289
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H et al (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14(9):865–868 (advance online publication)
Poirion OB, Zhu X, Ching T, Garmire LX (2017) Using single nucleotide variations in single-cell RNA-seq to identify tumor subpopulations and genotype-phenotype linkage. bioRxiv. https://www.biorxiv.org/content/early/2017/03/01/095810
Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P et al (2016) Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26(3):304–319
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A et al (2015) Tissue-based map of the human proteome. Science 347(6220):1260419–1260419
Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H et al (2017) A subcellular map of the human proteome. Science 356(6340):eaal3321
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1):W90–W97
Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM et al (2017) CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet 49(2):170–174
Xin J, Mark A, Afrasiabi C, Tsueng G, Juchler M, Gopal N et al (2016) High-performance web services for querying gene and variant annotation. Genome Biol 17(1):91
Jahn K, Kuipers J, Beerenwinkel N (2016) Tree inference for single-cell data. Genome Biol 17(1):86
Ross EM, Markowetz F (2016) OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol 17(1):69
Li H (2012) Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28(14):1838–1844
Campbell KR, Yau C, Rattray M (2016) Order Under Uncertainty: Robust Differential Expression Analysis Using Probabilistic Models for Pseudotime Inference. PLoS Comput Biol 12(11):e1005212
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21
Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C et al (2017) Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543(7643):122–125
Talevich E, Shain AH, Botton T, Bastian BC (2016) CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 12(4):e1004873
Silva GO, Siegel MB, Mose LE, Parker JS, Sun W, Perou CM et al (2017) SynthEx: a synthetic-normal-based DNA sequencing tool for copy number alteration detection and tumor heterogeneity profiling. Genome Biol 18(1):66
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499(7457):214–218
Leiserson MDM, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL et al (2014) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47(2):106–114
Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P et al (2016) Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 34(6):637–645
Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM et al (2017) Massively multiplex single-cell Hi-C. Nat Methods 14(3):263–266
Zhang J, Mardis ER, Maher CA (2017) INTEGRATE-neo: a pipeline for personalized gene fusion neoantigen discovery. Bioinformatics 33(4):555
Authors' contributions
LXG conceptualized the project. MAO wrote the manuscript, with assistance from OP, XZ, SH, TKW, RS and LXG. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank all the members of the Garmire Lab for their helpful discussions and manuscript review.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was supported by grants K01ES025434 awarded by NIEHS through funds provided by the trans-NIH Big Data to Knowledge (BD2 K) initiative (http://datascience.nih.gov/bd2k), P20 COBRE GM103457 awarded by NIH/NIGMS, R01 LM012373 awarded by NLM, and R01 HD084633 awarded by NICHD to LX Garmire.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ortega, M.A., Poirion, O., Zhu, X. et al. Using single-cell multiple omics approaches to resolve tumor heterogeneity. Clin Trans Med 6, 46 (2017). https://doi.org/10.1186/s40169-017-0177-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40169-017-0177-y