Abstract
Background
The gut microbiota is a reservoir of opportunistic pathogens that can cause life-threatening infections in critically ill patients during their stay in an intensive care unit (ICU). To suppress gut colonization with opportunistic pathogens, a prophylactic antibiotic regimen, termed “selective decontamination of the digestive tract” (SDD), is used in some countries where it improves clinical outcome in ICU patients. Yet, the impact of ICU hospitalization and SDD on the gut microbiota remains largely unknown. Here, we characterize the composition of the gut microbiota and its antimicrobial resistance genes (“the resistome”) of ICU patients during SDD and of healthy subjects.
Results
From ten patients that were acutely admitted to the ICU, 30 fecal samples were collected during ICU stay. Additionally, feces were collected from five of these patients after transfer to a medium-care ward and cessation of SDD. Feces from ten healthy subjects were collected twice, with a 1-year interval. Gut microbiota and resistome composition were determined using 16S rRNA gene phylogenetic profiling and nanolitre-scale quantitative PCRs.
The microbiota of the ICU patients differed from the microbiota of healthy subjects and was characterized by lower microbial diversity, decreased levels of Escherichia coli and of anaerobic Gram-positive, butyrate-producing bacteria of the Clostridium clusters IV and XIVa, and an increased abundance of Bacteroidetes and enterococci. Four resistance genes (aac(6′)-Ii, ermC, qacA, tetQ), providing resistance to aminoglycosides, macrolides, disinfectants, and tetracyclines, respectively, were significantly more abundant among ICU patients than in healthy subjects, while a chloramphenicol resistance gene (catA) and a tetracycline resistance gene (tetW) were more abundant in healthy subjects.
Conclusions
The gut microbiota of SDD-treated ICU patients deviated strongly from the gut microbiota of healthy subjects. The negative effects on the resistome were limited to selection for four resistance genes. While it was not possible to disentangle the effects of SDD from confounding variables in the patient cohort, our data suggest that the risks associated with ICU hospitalization and SDD on selection for antibiotic resistance are limited. However, we found evidence indicating that recolonization of the gut by antibiotic-resistant bacteria may occur upon ICU discharge and cessation of SDD.
Similar content being viewed by others
Background
The human gut microbiota comprises 1013–1014 bacterial cells that belong to hundreds of different species. The gut microbiota plays an important role in numerous metabolic, physiological, nutritional, and immunological processes of the human host [1]. In healthy individuals, the gut microbiota mostly consists of bacteria that have a commensal or mutualistic relationship with the human host. Critically ill patients, however, frequently have an extremely dysbiotic gut microbiota that is characterized by intestinal overgrowth with multi-drug resistant opportunistic pathogens of the phylum Proteobacteria (e.g., Escherichia coli) and the genus Enterococcus, while the abundance of commensal Bacteroidetes and Firmicutes is decreased [2,3,4,5]. The high levels of aerobic, opportunistic pathogens in the gut during critical illness are likely contributing to the burden of respiratory and bloodstream infections with these organisms in critically ill patients [6].
Selective digestive tract decontamination (SDD) aims to reduce the risk of nosocomial infections in ICU patients. SDD is used to eradicate opportunistic pathogens from the patients, while minimally impacting commensal bacteria [7]. In SDD, a paste containing the antibiotics colistin and tobramycin, and the antifungal amphotericin B, is applied to the oropharynx of ICU patients. The patients also receive a suspension of colistin, tobramycin, and amphotericin B via a nasogastric tube. These antimicrobials are applied from the day of ICU admission until ICU discharge. In addition, a third-generation cephalosporin (usually either cefotaxime or ceftriaxone) is administered intravenously during the first four days of ICU stay. SDD lowers patient mortality during ICU stay in settings with a low prevalence of antibiotic resistance and reduces the costs associated with ICU hospitalization [8, 9]. Selection of bacteria that are resistant to the antimicrobials used in SDD remains a major concern [10, 11], although this is not supported by the results of clinical trials in which conventional culture techniques were used to screen for antibiotic resistance among nosocomial pathogens [12].
The patient gut not only is a potential source for opportunistic pathogens but also forms a large reservoir for antibiotic resistance genes, termed the gut resistome [13,14,15,16,17]. The use of antibiotics may favor the selection for antimicrobial resistance genes (ARGs) among members of the gut microbiota, thus increasing the likelihood of horizontal spread of ARGs between commensals and opportunistic pathogens co-residing in the gut [16]. During the administration of SDD, the gut resistome of patients is monitored by the cultivation of resistant bacteria from rectal swabs or feces, as part of routine diagnostics. However, methods that rely on microbial culture capture only a fraction of the gut resistome, since anaerobic commensals, which are the main reservoir of ARGs in the gut microbiota, are difficult to culture [18,19,20]. Thus, culture-independent methods are needed to comprehensively assess the impact of antibiotic prophylaxis on the microbiota and resistome of ICU patients.
Here, we used the 16S ribosomal RNA (rRNA) gene-targeted Human Intestinal Tract Chip (HITChip) and nanolitre-scale quantitative PCR (qPCR) targeting 81 ARGs to determine the dynamics of gut microbiota composition and resistome in patients receiving SDD during ICU hospitalization. We contrast these findings in ICU patients with the composition of the microbiota and resistome of healthy subjects.
Methods
Study population
All included patients (n = 10) were acutely admitted to the ICU of the University Medical Center Utrecht from the community and had not been hospitalized in the previous six months, with the exception of patient 105 who was hospitalized for five days prior to transfer to the ICU and start of SDD. None of the patients were treated with antibiotics in six months prior to ICU hospitalization. All patients received SDD from the start of ICU stay until ICU discharge. SDD consists of 1000 mg of cefotaxime intravenously four times daily for four days; an oropharyngeal paste containing polymyxin E, tobramycin, and amphotericin B (each in a 2% concentration); and administration of a 10 mL suspension containing 100 mg polymyxin E, 80 mg tobramycin, and 500 mg amphotericin B via a nasogastric tube, four to eight times daily throughout ICU stay. All patients received additional antibiotics during ICU stay. Fecal samples of patients were collected by nursing staff upon defecation and stored at 4 °C for 30 min to 4 h, after which the samples were transferred to −80 °C. Seven patients included here (patient numbers 105, 108, 120, 163, 164, 165, and 169) were also included in a previous study where the dynamics of two aminoglycoside resistance genes in the gut microbiota of ICU patients was studied [20]. A total of 30 fecal samples were collected during ICU stay. Five additional fecal samples were collected after transfer to a medium-care ward and cessation of SDD. Additional file 1 includes detailed information on sampling time points and antibiotic usage of the ICU patients in this study.
Routine surveillance for colonization with aerobic Gram-negative bacteria in ICU patients was performed through culturing of rectal swabs on sheep blood agar and MacConkey agar. All suspected Gram-negative colonies were analyzed by Maldi-TOF for species identification. Antibiotic resistance phenotypes were determined using the Phoenix system (BD, Franklin Lakes, NJ, USA).
Fecal samples of healthy subjects were collected as part of the “Cohort study of intestinal microbiome among Irritable Bowel Syndrome patients and healthy individuals” (CO-MIC) study at two time points with a one-year interval between sampling. None of the individuals in this cohort received antibiotics. All included patients and healthy subjects were adults. Further demographic information on both cohorts is provided in Additional file 2. DNA from fecal samples of patients and healthy subject was isolated as previously described [21], using two rounds of bead beating and purification using the QIAamp DNA Stool Mini Kit columns (Qiagen; Venlo, The Netherlands).
Gut microbiota profiling by HITChip
The HITChip is a validated phylogenetic array produced by Agilent Technologies (Palo Alto, CA) and developed at Wageningen University, The Netherlands [22, 23]. It contains over 4800 oligonucleotides targeting the V1 and the V6 region of the 16S rRNA gene from 1132 microbial phylotypes present in the human gut [22]. The full-length 16S rRNA gene was amplified from fecal DNA, and PCR products were further processed and hybridized to the microarrays as described previously [24]. Data analyses were performed using R (www.r-project.org), including the microbiome package (https://github.com/microbiome). Bacterial associations in the different patient groups and healthy subjects were assessed using principal component analysis (PCA) as implemented in CANOCO 5.0 [25]. Redundancy analysis (RDA) was performed as implemented in CANOCO 5.0 to determine the associations between microbiota composition (based on 130 genus-like groups included in the HITChip) and explanatory host-associated variables (age, gender, body mass index (BMI), and sample source, i.e., ICU patient or healthy subject). Significance of the explanatory variables was assessed by Monte Carlo permutation testing (MCPT). Statistical testing for the differences in microbiota composition between ICU patients and healthy subjects was performed by the non-parametric Mann-Whitney U test. All P values were corrected for false discovery rate (FDR) by the Benjamini and Hochberg method [26], and corrected P values (q) below 0.05 were considered significant.
Quantification of E. coli by qPCR
qPCRs for the quantification of E. coli were performed with primers that were previously described [27], using serial dilutions of genomic DNA of E. coli DH5α to generate a standard curve. The quantification of 16S rRNA was performed with primers described in [28]. The PCR conditions were identical to the qPCR conditions for the detection of mcr-1 (described below).
qPCR analysis of antibiotic resistance genes
qPCR analysis was performed using the 96.96 BioMark™ Dynamic Array for Real-Time PCR (Fluidigm Corporation, San Francisco, CA), according to the manufacturer’s instructions, with the exception that the annealing step in the PCR was held at 56 °C. Fecal DNA was first subjected to 14 cycles of specific target amplification using a 0.18-μM mixture of all primer sets, excluding the 16S rRNA primer sets, in combination with the Taqman PreAmp Master Mix (Applied Biosystems), followed by a fivefold dilution prior to loading samples onto the Biomark array for qPCR. Thermal cycling and real-time imaging was performed on the BioMark instrument, and Ct values were extracted using the BioMark Real-Time PCR analysis software.
Target selection, primer design, and primer validation
The primer set used in the qPCR assays covered 81 antimicrobial resistance genes (ARGs) of 14 resistance gene classes (Additional file 3). Primers were designed for the ARGs that are most commonly detected in the gut microbiota of healthy individuals [14, 15] and clinically relevant ARGs, including genes encoding extended spectrum β-lactamases (ESBLs), carbapenemases, and proteins involved in vancomycin resistance. Primer design was performed using Primer3 [29] with its standard settings with a product size of 80–120 bp and a primer melting temperature of 60 °C. The universal primers for 16S rRNA genes were previously described by Gloor et al. [28]. Forward and reverse primers were evaluated in silico for cross hybridization using BLAST [30] and were cross-referenced against ResFinder [31] to ensure the correct identity of the targeted genes. All primers that aligned with more than ten nucleotides at their 3′ end to another primer sequence were discarded and redesigned. Additionally, all primer sets were aligned to all resistance genes that were targeted in this PCR analysis to test for cross hybridization with genes other than the intended target resistance gene. Primers that aligned with more than ten nucleotides at their 3′ end sequence with a non-target resistance gene were discarded and redesigned. A reference sample consisting of pooled fecal DNA from different patients was loaded in a series of fourfold dilutions and was used for the calculation of primer efficiency. All primers whose efficiency was experimentally determined to be between 80 and 120% were used to determine the normalized abundance of the target genes. The detection limit on the Biomark system was set to a CT value of 20, as recommended by the manufacturer. In addition, to assess primer specificity, we performed melt curve analysis using the Fluidigm melting curve analysis software (http://fluidigm-melting-curve-analysis.software.informer.com/). All PCRs were performed in triplicate, and sample-primer combinations were only included in the analysis when all triplicate reactions resulted in a CT value below the detection limit.
After completion of the nanolitre-scale qPCR assays, the transferable colistin resistance gene mcr-1 was described [32]. To detect and quantify mcr-1, we developed primers (qPCR-mcr1-F: 5′-TCGGACTCAAAAGGCGTGAT-3′ and qPCR-mcr1-R: 5′-GACATCGCGGCATTCGTTAT-3′) for use in a standard qPCR assay. The mcr-1 gene was synthesized based on the sequence described in [32] by Integrated DNA Technologies (Leuven, Belgium) and used as a positive control in our assays. The qPCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific, Leusden, The Netherlands) and a StepOnePlus instrument (Applied Biosystems, Nieuwekerk a/d IJssel, The Netherlands) with 5 ng DNA in the reaction and the following program: 95 °C for 10 min and subsequently 40 cycles of 95 °C for 15 s and 56 °C for 1 min.
Calculation of normalized and cumulative abundance
Normalized abundance of resistance genes was calculated relative to the abundance of the 16S rRNA gene (CTARG − CT16S rRNA), resulting in a log2-transformed estimate of ARG abundance. Statistical analysis was performed using GraphPad Prism (La Jolla, CA). Visualization of the qPCR data in the form of a heat map was performed using Microsoft Excel. Statistical testing of the differences in the abundance of resistance genes in ICU patients versus healthy subjects was performed with the non-parametric Mann-Whitney U test with a false discovery rate (Benjamini Hochberg) <0.05 to correct for multiple testing.
Results
Microbiota dynamics in ICU patients and healthy subjects
Ten ICU patients and ten healthy subjects were included in this study. There were no statistically significant differences between the cohorts in their gender distribution and BMI. The age of the ICU patients was significantly (P < 0.05; Mann-Whitney U test) higher (median 57, interquartile range 49–74) than the age of the healthy subjects (median 42.5, interquartile range 31–51; Additional file 2). Global changes in the gut microbiota of healthy subjects and ICU patients were visualized by principal component analysis (Fig. 1a). The microbiota profiles of healthy subjects clustered together, indicating that they had stable and broadly comparable microbiota profiles, which were clearly distinct from the microbiota profiles of patients during and after ICU stay. These profiles covered a larger area in the PCA plot, indicating that the differences in the microbiota composition of patients were larger than that of healthy subjects. The composition of the gut microbiota in patient samples collected during and after ICU hospitalization was markedly altered, with samples clustering away from the healthy controls. RDA showed that both sample source (i.e., whether samples were taken from ICU patients or healthy subjects) and, to a lesser extent, BMI significantly explained microbiota composition variance (sample source: q = 0.007; BMI: q = 0.048, Additional file 4). The other tested variables (age and gender) did not significantly contribute to the variability in microbiota composition. Collectively, the tested variables explained 30% of microbiota composition variance.
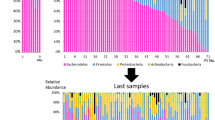
Dynamics of gut microbiota composition and diversity in ICU patients and healthy subjects. a Principal component analysis (PCA) of gut microbiota composition of ICU patients. Dashed symbols indicate fecal samples collected after ICU discharge and continued hospitalization in a medium-care ward. Fecal samples that were collected in the first 5 days of ICU hospitalization are indicated by a black line around the symbol. Fecal samples of healthy subjects were collected at two time points with 1-year interval, indicated with black and gray circles, respectively. b Diversity (Shannon index) of the microbiota of ICU patients. Double lines indicate hospitalization in a medium-care ward. c Diversity (Shannon index) of the microbiota of healthy subjects. d Gut microbiota composition of patients and healthy subjects. Stacked bar charts represent the abundance of different major taxa in the gut microbiota of ICU patients and healthy subjects. Among Bacilli, the genus Enterococcus has been highlighted, as SDD has previously been shown to select for colonization with enterococci [36, 37]. Fecal samples that were collected after ICU discharge and during medium-care hospitalization are indicated by gray triangles. Statistically significant differences of the abundance of taxa in the gut microbiota of patients during ICU hospitalization and healthy subjects are indicated in the legend (*q < 0.05; **q < 0.01; ***q < 0.001; Mann-Whitney U test with Benjamini-Hochberg correction for multiple testing)
The diversity of the microbiota, as quantified by Shannon’s diversity index, was significantly lower in ICU patients compared to healthy subjects (5.90 ± 0.20 vs 5.19 ± 0.46, respectively; P < 0.001, Student’s t test). The diversity of the microbiota of ICU patients was highly dynamic (Fig. 1b). Several patients (#108, #163, #164, #165, and #169) experienced a rapid loss of diversity in the first days of ICU stay. In contrast, the diversity of the microbiota was more stable in healthy subjects when comparing samples that were collected 1 year apart (Fig. 1c). Compared to healthy subjects, the microbiota of patients during ICU hospitalization was characterized by a significantly higher abundance in the taxa Bacteroidetes and Bacilli: Enterococcus and a lower abundance of the taxa Clostridium cluster IV and XIVa (Fig. 1d).
We performed quantitative PCRs to accurately determine the abundance of E. coli, one of the primary targets of SDD, in the gut microbiota of patients and healthy subjects (Fig. 2). The abundance of E. coli in samples of ICU patients was lower compared to the healthy subjects (p = 0.001; Mann-Whitney U test). Notably, upon ICU discharge, cessation of SDD, and transfer to a medium-care ward, the abundance of E. coli rebounded in one patient (#105) to levels surpassing those found in healthy individuals.
Abundance of E. coli in the gut microbiota of ICU patients and healthy subjects. Quantification of E. coli 16S rRNA gene copies relative to total 16S rRNA gene copies, performed by qPCR with three technical replicates. Error bars indicate standard deviation. Samples are ordered by time of sampling during ICU stay. The color coding of the samples is unique for each patient and is identical to Fig. 1. Statistical testing was performed with the Mann-Whitney U test
During ICU stay, routine surveillance by conventional microbiological culture was performed on all patients. E. coli could be cultured from six out of 73 rectal swabs that were collected during the patients’ ICU stay. Five E. coli-positive rectal swabs, of patients #43, #105, #108, #163, and #169, were collected within one day of ICU admission, while the sixth positive swab (of patient #165) was collected after 9 days of ICU stay. In addition, an E. coli strain from patient #105 with an ESBL-producing and tobramycin-resistant phenotype was isolated after ICU discharge, while the patient was in a medium-care ward. The E. coli strains isolated during ICU stay were susceptible to cephalosporins and aminoglycosides. All E. coli strains were susceptible to colistin.
Resistome dynamics in ICU patients and healthy subjects
A total of 46 unique ARGs conferring resistance to 12 different classes of antimicrobials were detected in the DNA isolated from fecal samples of hospitalized patients and healthy subjects (Additional file 5). The number of detected resistance genes per sample ranged between 6 and 38. Eleven resistance genes were detected in >80% of healthy subjects and critically ill patients. This highly prevalent set of resistance genes included tetracycline resistance genes (tetO, tetQ, tetM, tetW), two aminoglycoside resistance genes (aph(3′)-III and an aadE-like gene), the bacteroidal β-lactam resistance gene cblA, and the macrolide resistance gene ermB.
Genes associated with major antibiotic resistance threats, including those identified by the Centers for Disease Control, were relatively rare. Genes encoding for extended-spectrum beta-lactamases (ESBLs) were not detected in healthy subjects. In two ICU patient samples (#105C and #108C), however, the ESBL genes bla CTX-M and bla DHA , respectively, could be detected. Sample #105C was collected after ICU discharge and cessation of SDD, while sample #108C was collected after 12 days of ICU hospitalization and SDD treatment. The carbapenemase bla KPC was detected in a single patient (patient #180), but only in the first sample (collected after five days in the ICU) and not in the second sample, which was collected after 16 days of ICU hospitalization. No other ESBL- or carbapenemase-producing strains were isolated from the patients during ICU hospitalization. Other enterobacterial β-lactamases were found to be widespread in our resistome analysis. The bla AMPC β-lactamase was present in 37% of samples, with nine of ten patients and eight of ten healthy subjects having detectable levels of bla AMPC at one or more sampling points. The bla TEM β-lactamase was present in 26% of samples, corresponding with five of ten patients and four of ten healthy subjects in which this gene was detectable at one or more sampling points. None of the samples were positive for the carbapenemases bla NDM and bla OXA or the transferable colistin resistance gene mcr-1. Among resistance genes that are associated with Gram-positive pathogens, the staphylococcal methicillin resistance gene mecA was detected in 13 samples from eight of ten patients, but not in samples of healthy subjects. The vancomycin resistance gene vanB was present in five samples from three of ten patients and six samples from four of ten healthy subjects.
A comparison of the abundance of individual ARGs in samples that were collected during ICU stay, versus samples from healthy subjects, revealed that four ARGs (aac(6′)-Ii, ermC, qacA, tetQ) were significantly more abundant in ICU patients, while two ARGs (catA and tetW) were significantly more abundant in healthy individuals (Fig. 3).
Antimicrobial resistance genes present at significantly higher or lower levels in the microbiota of ICU patients, compared to healthy subjects. ARGs that are present at significantly higher (aac(6′)-Ii, ermC, qacA, and tetQ) or lower (catA and tetW) abundance in ICU patients, compared to healthy subjects, are shown. Testing for statistically significant differences was performed by the Mann-Whitney U test, with Benjamini-Hochberg correction for multiple testing (*q < 0.05; **q < 0.01). The horizontal line denotes the median value. The detection limit of the qPCR assay is indicated with the dashed line
Discussion
Current guidelines in The Netherlands recommend topical antibiotic decontamination in ICU patients with an expected ICU stay of two days or longer. Yet, the original claim that these interventions do not affect harmless anaerobic intestinal bacteria [7] has recently been questioned [20, 33]. While culture-based studies did not demonstrate selection for antibiotic-resistant opportunistic pathogens during SDD treatment [9, 12, 34, 35], concerns remain that selection for antibiotic resistance genes occurs in the gut microbiota of patients that are treated by SDD during their stay in the ICU.
The current study describes the composition of the gut microbiota and the resistome of ICU patients receiving SDD during ICU stay and compares these findings to the microbiota and resistome of healthy subjects. We were not able to include an ICU control group that was not treated with SDD, as this would be a breach of clinical guidelines for ICU patients in our country. For this reason, it is not possible to disentangle the effects of SDD from other factors that affect the composition of the microbiota during ICU stay, including underlying critical illness, parenteral feeding, and curative antibiotic therapy.
The gut microbiota of ICU patients in this study was characterized by a low diversity, the increased abundance of enterococci, and lower abundance of anaerobic Gram-positive, butyrate-producing bacteria of the Clostridium clusters IV and XIVa. These findings are in line with previous studies reporting selection for Gram-positive cocci [12, 20, 36, 37] and depletion of Faecalibacterium prausnitzii in ICU patients receiving SDD [33]. In addition, we were able to demonstrate that the abundance of E. coli was significantly lower in ICU patients than in healthy individuals. The suppression of E. coli in the SDD-treated ICU patients starkly contrasts with other studies in critically ill patients not receiving SDD, in which E. coli is present at higher levels than in healthy individuals [2, 3, 5]. This observation further supports previous studies which found that SDD is successful in suppressing outgrowth of E. coli in the gut microbiota of ICU patients [8, 9], corresponding to the original aim of SDD [7].
Notably, levels of E. coli increased again after ICU discharge in two of patients, reaching levels in the gut similar to, or even surpassing, those in healthy individuals. These findings suggest that a rapid regrowth or recolonization of the intestinal tract by E. coli, and possibly other aerobic Gram-negative bacteria, occurs upon cessation of prophylactic antibiotic therapy. In one of the post-ICU samples, an ESBL-producing E. coli strain was isolated. Indeed, this was the only sample in which the ESBL bla CTX-M gene was detected in our resistome analysis. In the only prospective evaluation on the post-ICU effects of SDD, the implementation of SDD was not associated with higher infection rates after ICU discharge [38], but further studies are warranted to better quantify the risks associated with recolonization of the gut by multi-drug resistant, opportunistic pathogens upon ICU discharge, and cessation of SDD.
The qPCR-based analysis of the resistome confirms previous metagenomic studies, in showing that tetracycline and aminoglycoside resistance genes and bacteroidal β-lactamases are widespread in the human intestinal microbiota [14, 15, 18, 20]. Four resistance genes (aac(6′)-Ii, ermC, qacA, tetQ) were significantly more abundant among ICU patients than in healthy subjects. The aac(6′)-Ii gene is a specific chromosomal marker for the nosocomial pathogen Enterococcus faecium and provides low-level resistance to aminoglycosides [39]. Its high abundance in ICU patients is in line with the increased levels of enterococci in the microbiota of the patients. The increased abundance of the macrolide resistance gene ermC may have been selected for by the use of low doses of the macrolide erythromycin, which was used as an agent to accelerate gastric emptying during ICU stay in six patients. The increased abundance of tetQ in the gut microbiota of ICU patients may reflect the higher abundance of Bacteroidetes in ICU patients versus healthy subjects, as tetQ is widely distributed on conjugative transposons in this phylum [40]. Finally, the qacA gene confers resistance to a number of disinfectants, including the biguanidine compound chlorhexidine and the quaternary ammonium compound benzalkonium chloride [41, 42]. Disinfectants are widely used in ICUs as cleaning and infection control agents [43], and their use could select for qacA in the gut microbiota of patients.
Two resistance genes (catA and tetW) were more abundant in healthy individuals than in ICU patients. There is currently little information on the distribution of the catA gene among bacteria associated with the human gut microbiota, but the gene was frequently found in human feces in a recent study set in low-income human habitats [44]. The tetracycline resistance gene tetW is present in Gram-positive anaerobic gut commensals [45].
Although SDD improves survival of ICU patients, its use remains controversial due to the risk for selection of antibiotic resistance among bacteria that populate the patient gut. In this observational study, we found little evidence for the selection of high-risk resistance determinants, like ESBLs, carbapenemases, or vancomycin resistance genes, in SDD-treated patients during their stay in the ICU. The increased abundance of the resistance genes aac(6′)-Ii, ermC, qacA, and tetQ in SDD-treated ICU patients in our study could be interpreted as being of limited concern. The first three resistance genes contribute to resistance in enterococci, either to relatively low concentrations of antibiotics (aac(6′)-Ii) or to classes of antimicrobials that are of limited relevance for the treatment of enterococcal infections (ermC and qacA). The tetQ gene provides resistance to tetracyclines in Bacteroidetes, but this class of antibiotics is scarcely used for the treatment of anaerobic infections [46]. The selection for enterococci occurring in this patient cohort may be of concern in settings where vancomycin-resistant enterococci (VRE) are endemic. VRE is still infrequent in The Netherlands, and the vanB resistance gene that was detected in both patients and healthy subjects is most likely associated with Gram-positive anaerobes [47]. It remains to be determined whether SDD leads to selection for VRE in countries where these bacteria are more prevalent.
Conclusions
Our data support the notion that in settings with low levels of circulating antibiotic resistance genes, ICU hospitalization and SDD treatment does not lead to the selection of antibiotic resistance genes with high clinical relevance. However, it is difficult to generalize these findings from a Dutch hospital to other countries, where resistant bacteria are more prevalent. Our study also illustrates the relative lack of data regarding recolonization of the gut upon ICU discharge and cessation of SDD. Future studies into the emergence and spread of antibiotic resistance genes in ICU patients can benefit from the qPCR platform used here as it enables the rapid detection and quantification of antibiotic resistance genes in the gut microbiota. The detection of high-risk antibiotic resistance genes (encoding, e.g., ESBLs, carbapenemases or vancomycin resistance proteins) in the resistome of patients may lead to the implementation of targeted antibiotic therapy or infection control measures to minimize the risk for selection and spread of these resistance genes.
References
Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904.
Ojima M, Motooka D, Shimizu K, Gotoh K, Shintani A, Yoshiya K, et al. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci. 2016;61:1628–34.
McDonald D, Ackermann G, Khailova L, Baird C, Heyland D, Kozar R, et al. Extreme dysbiosis of the microbiome in critical illness. mSphere. 2016;1:e00199–16.
Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio. 2014;5:e01361–14.
Yeh A, Rogers MB, Firek B, Neal MD, Zuckerbraun BS, Morowitz MJ. Dysbiosis across multiple body sites in critically ill adult surgical patients. Shock. 2016;46(6):649–54.
Donskey CJ. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis. 2004;39:219–26.
van der Waaij D, Manson WL, Arends JP, de Vries-Hospers HG. Clinical use of selective decontamination: the concept. Intensive Care Med. 1990;16(Suppl 3):S212–6.
de Jonge E, Schultz MJ, Spanjaard L, Bossuyt PMM, Vroom MB, Dankert J, et al. Effects of selective decontamination of digestive tract on mortality and acquisition of resistant bacteria in intensive care: a randomised controlled trial. Lancet. 2003;362:1011–6.
de Smet AMGA, Kluytmans J a JW, Cooper BS, Mascini EM, RFJ B, van der Werf TS, et al. Decontamination of the digestive tract and oropharynx in ICU patients. N Engl J Med. 2009;360:20–31.
Philips BJ. Selective decontamination of the digestive tract: time to implement it in all UK intensive care units? Maybe not yet. Br J Anaesth. 2014;113:537–9.
Wunderink RG. Welkommen to our world. Emergence of antibiotic resistance with selective decontamination of the digestive tract. Am J Respir Crit Care Med. 2010;181:426–7.
Daneman N, Sarwar S, Fowler RA, Cuthbertson BH, SuDDICU Canadian Study Group. Effect of selective decontamination on antimicrobial resistance in intensive care units: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13:328–41.
van Schaik W. The human gut resistome. Philos Trans R Soc Lond Ser B Biol Sci. 2015;370:20140087.
Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A, et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–9.
Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4:2151.
Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–6.
Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4:54.
Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Buelow E, Gonzalez TB, Versluis D, Oostdijk EAN, Ogilvie LA, van Mourik MSM, et al. Effects of selective digestive decontamination (SDD) on the gut resistome. J Antimicrob Chemother. 2014;69:2215–23.
Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–34.
Rajilić-Stojanović M, Heilig HGHJ, Molenaar D, Kajander K, Surakka A, Smidt H, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–51.
van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–80.
Jalanka-Tuovinen J, Salonen A, Nikkilä J, Immonen O, Kekkonen R, Lahti L, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One. 2011;6:e23035.
Ter Braak CJF, Smilauer P. Canoco reference manual and user’s guide: software for ordination. Ithaca: Microcomput. Power; 2012.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300.
Furet J-P, Firmesse O, Gourmelon M, Bridonneau C, Tap J, Mondot S, et al. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol. 2009;68:351–62.
Gloor GB, Hummelen R, Macklaim JM, Dickson RJ, Fernandes AD, MacPhee R, et al. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One. 2010;5:e15406.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4.
Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8.
Benus RF, Harmsen HJ, Welling GW, Spanjersberg R, Zijlstra JG, Degener JE, et al. Impact of digestive and oropharyngeal decontamination on the intestinal microbiota in ICU patients. Intensive Care Med. 2010;36:1394–402.
Oostdijk EN, Kesecioglu J, Schultz MJ, et al. Effects of decontamination of the oropharynx and intestinal tract on antibiotic resistance in ICUs: a randomized clinical trial. JAMA. 2014;312:1429–37.
Plantinga NL, Bonten MJ. Selective decontamination and antibiotic resistance in ICUs. Crit Care. 2015;19:259.
van der Bij AK, Frentz D, Bonten MJM, ISIS-AR Study Group. Gram-positive cocci in Dutch ICUs with and without selective decontamination of the oropharyngeal and digestive tract: a retrospective database analysis. J Antimicrob Chemother. 2016;71:816–20.
Humphreys H, Winter R, Pick A. The effect of selective decontamination of the digestive tract on gastrointestinal enterococcal colonization in ITU patients. Intensive Care Med. 1992;18:459–63.
de Smet AMGA, Hopmans TEM, Minderhoud ALC, Blok HEM, Gossink-Franssen A, Bernards AT, et al. Decontamination of the digestive tract and oropharynx: hospital acquired infections after discharge from the intensive care unit. Intensive Care Med. 2009;35:1609–13.
Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37:1896–903.
Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ Microbiol. 2001;67:561–8.
Mitchell BA, Brown MH, Skurray RA. QacA multidrug efflux pump from Staphylococcus aureus: comparative analysis of resistance to diamidines, biguanidines, and guanylhydrazones. Antimicrob Agents Chemother. 1998;42:475–7.
Tennent JM, Lyon BR, Midgley M, Jones G, Purewal AS, Skurray RA. Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Microbiology. 1989;135:1–10.
McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–79.
Pehrsson EC, Tsukayama P, Patel S, Mejía-Bautista M, Sosa-Soto G, Navarrete KM, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–6.
Scott KP, Melville CM, Barbosa TM, Flint HJ. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother. 2000;44:775–7.
Brook I. Treatment of anaerobic infection. Expert Rev Anti-Infect Ther. 2007;5:991–1006.
Graham M, Ballard SA, Grabsch EA, Johnson PDR, Grayson ML. High rates of fecal carriage of nonenterococcal vanB in both children and adults. Antimicrob Agents Chemother. 2008;52:1195–7.
Acknowledgements
We thank ServiceXS B.V. (Leiden, The Netherlands) for their assistance in the Fluidigm real-time PCR assays. We are grateful to Erwin Zoetendal and Willem M. de Vos for providing the material and data from the Cohort study of intestinal microbiota among irritable bowel syndrome patients and healthy individuals’ (CO-MIC) funded by the unrestricted Spinoza Award to Willem M. de Vos from The Netherlands Organization for Scientific Research.
Funding
This work was supported by The Netherlands Organisation for Health Research and Development ZonMw (Priority Medicine Antimicrobial Resistance; grant 205100015) and by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) “Evolution and Transfer of Antibiotic Resistance” (EvoTAR), under grant agreement number 282004. In addition, W.v.S is supported by a NWO-VIDI grant (917.13.357) and L.L. by Academy of Finland grants 295741 and 307127.
Availability of data and materials
The 16S rRNA gene profiling datasets generated in the current study are available in the Figshare repository (https://doi.org/10.6084/m9.figshare.5116795). Other generated and analyzed (meta)data are included in this publication.
Author information
Authors and Affiliations
Contributions
RJLW, MJMB, MWJvP, HS, and WvS designed the study. EB, MSvM, EANO, MWJvP, HS, and WvS supervised the collection of fecal samples. EB, TdJBG, EAMM, and JCB performed the experiments to map the gut resistome and phylogenetic composition of the microbiome. SF, WAAdSP, LL, and JRB contributed to the bioinformatic analyses. The manuscript was written by EB, TdJBG, SF, RJLW, MJMB, MWJvP, HS, and WvS and approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol for the ICU patient arm of this study was reviewed and approved by the institutional review board of the University Medical Center Utrecht (Utrecht, The Netherlands) under number 10/0225. Informed consent for fecal sampling during hospitalization was waived. The protocol for the feces collection of healthy subjects, including informed consent, was reviewed and approved by the Ethics Committee of Gelderse Vallei Hospital (Ede, The Netherlands).
Consent for publication
Not applicable.
Competing interests
W.v.S. has served as a consultant for Vedanta Biosciences. The other authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Patient details. The antibiotics used in treatment of patients during hospitalization and time points at which fecal samples were collected are indicated. SDD indicates the administration of topical components of SDD, i.e., a paste containing polymyxin E, torbramycin, and amphotericin B (each at 2%) applied to the oropharynx and the administration of a 10 mL suspension containing 100 mg polymyxin E, 80 mg tobramycin, and 500 mg amphothericin B via nasogastric tube. Black lines indicate hospitalization at the ICU, and blue lines indicate hospitalization at a medium-care ward. (PDF 309 kb)
Additional file 2:
Demographic details of patients and healthy subjects. Gender, age, weight, and body mass index (BMI) are indicated. The statistical analysis of differences between the two cohorts was performed with the χ2 test (for gender) and Mann-Whitney U test (for age, weight, and BMI). ND: not determined. (XLSX 11 kb)
Additional file 3:
Primers used in this study. Primers were developed to target the indicated ARGs. Primer sequences in bold indicate ARGs which were detected in ≥1 sample. (DOCX 39 kb)
Additional file 4:
Redundancy analysis (RDA) of variance in microbiota composition. RDA was performed as implemented in CANOCO 5.0 [25] to determine the associations between microbiota composition and explanatory host-associated variables. Significance of the explanatory variables was assessed by MCPT. Statistical testing for the differences in microbiota composition between ICU patients and healthy subjects was performed by the Mann-Whitney U test with FDR correction by the Benjamini and Hochberg method. (XLSX 9 kb)
Additional file 5:
Dynamics of the resistome in ICU patients and healthy subjects. ARGs are grouped and color-coded according to resistance gene families (B: bacitracin, C: chloramphenicol; M: macrolides; P, polymyxins; Qa: quaternary ammonium compounds, S: sulphonamides; Tet: tetracyclines; T: trimethoprim; V: vancomycin). Abundance (log2-transformed) is visualized relative to 16S rRNA. The time points at which samples were collected during hospitalization are indicated. Patients are color-coded as in Fig. 1. Samples indicated with a lighter color have been collected after cessation of SDD during medium-care hospitalization. Red and green boxes indicate antibiotic resistance genes that were significantly (q < 0.05, Mann-Whitney test with Benjamini-Hochberg correction) more or less abundant, respectively, in the gut microbiota of ICU patients compared to healthy subjects. (PDF 331 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Buelow, E., Bello González, T.d.j., Fuentes, S. et al. Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome 5, 88 (2017). https://doi.org/10.1186/s40168-017-0309-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40168-017-0309-z