Abstract
Background
The human concentrative nucleoside transporter 1 (hCNT1) a product of the SLC28A1 gene is one of the three concentrative nucleoside transporters, with a substrate specificity for physiological pyrimidine nucleosides. It has recently been implicated in tumor suppression. We have unraveled a splice variant RNA transcript that is overexpressed in some tumor tissues and some cancer cells. This study established that observation.
Methods
We examined several clones of hCNT1 generated from RT-PCR of total RNA from human kidney tissue purchased from Ambion. The resulting cDNA clones were then sequenced, and a variant that retained intron 4, and skipped some exons fully or partly, specifically exons 5 and 13 were completely missed and only part of exon 6 was spliced. Tissue expression analysis by PCR indicated a similar distribution of expression of RNA of the splice variant hCNT1-IR as that of the dominant variant hCNT1, particularly in the small intestine, kidney and liver. Further, analysis of various tumor samples with PCR primers designed from this novel hCNT1 splice variant (hCNT1-IR) revealed interestingly that it is overexpressed in some cancer tissues relative to normal tissues, particularly kidney, liver and pancreatic cancers.
Conclusion
We have identified a novel intron retaining and exon skipping splice variant of the hCNT1 nucleoside transporter, and designated it hCNT1-IR, which has a similar tissue expression distribution as the normal hCNT1 variant, but unlike the normal transcript, hCNT1-IR is overexpressed in some cancers and may serve as a potential cancer biomarker.
Similar content being viewed by others
Introduction
Physiological nucleosides and most therapeutic nucleoside analogues are hydrophilic molecules that require specialized nucleoside transporter (NT) proteins to pass across cellular membranes. NT-mediated transport is a critical determinant of intracellular nucleoside metabolism and the pharmacological actions of antineoplastic and antiviral nucleoside drugs [1, 2]. Nucleoside transporters are divided into two families: the SLC28 family comprising concentrative nucleoside transporters (CNTs) and the SLC29 family comprising equilibrative nucleoside transporters (ENTs). CNT1 is the first member of the CNT family that was cloned and functional characterized, and is predominantly expressed in kidney and jejunum [3]. CNT1 mediates the cellular uptake of naturally occurring pyrimidine nucleosides and adenosine to a much lesser extent, as well as diverse anticancer and antiviral nucleoside analogues including gemcitabine, cytarabine, and zidovudine [4,5,6,7].
CNT1 is a highly variable gene. A number of coding region single nucleotide polymorphisms (SNPs) have been reported for human CNT1 (see http://www.pharmgkb.org and http://www.pharmacogenetics.ucsf.edu). Sixty variable sites in the coding and flanking intronic regions of hCNT1 were identified in 247 DNA samples from an ethnically diverse population. Insertion (hCNT1 + Val) and deletion (hCNT1-1153del) mutations were also identified [8]. Initial cloning of human CNT1 from kidney yielded three highly similar clones, hCNT1a, hCNT1b, and hCNT1c, which may represent genetic variants of human CNT1 [3]. Coding region SNPs have also been reported for CNT1 by a study in a Japanese population [9]. The SNP database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP) from that study lists the non-synonymous and synonymous changes for CNT1. In addition, one study has identified a cDNA variant of CNT1 with an additional 116 bp in the 5′-untranslated region from a human fetal liver cDNA library [10]. Gray et al. have studied the function of single amino acid variants of human CNT1 as a starting point in the analysis of genetic influence on variation in transporter function [11]. Variation in splice sites and promoter regions may also influence transporter expression or function. Hence, determining the functional roles of genetic and splice variants of concentrative nucleoside transporters in the targeting and disposition of naturally occurring nucleosides and synthetic nucleoside analogs is an important focus for future studies.
Alternative splicing is a very frequent phenomenon in the human transcriptome. There are four major types of alternative splicing: exon skipping, alternative 3′ splice site, alternative 5′ splice site, and intron retention [12]. Such variation can be functional or can represent mis-splicing. Alternative splicing can either produce functional alternative transcripts with distinct functions or might modulate functional spliced transcript level by producing some transcripts that do not encode proteins (sometimes by leaving some introns unspliced [13]. Intron retention, one form of alternative splicing, is common in plants but rare in higher eukaryotes, because messenger RNAs with retained introns are subject to cellular restriction at the level of cytoplasmic export and expression. In humans, 2–5 % of the genes have been reported to appear in the form of intron retentions [14, 15]. A recent study showed a splice variant of the organic cation transporter OCTN2 in which a 72-base-pair sequence located in the first intron of OCTN2 gene was spliced between exons 1 and 2, was retained in the endoplasmic reticulum, and did not transport carnitine [16]. Alternative pre-mRNA splicing increases proteomic diversity and provides a potential mechanism underlying both phenotypic diversity and susceptibility to genetic disorders in human populations. In the present study, we cloned a novel splice variant of hCNT1 with intron 4 retention from human kidney. The expression of protein and function of the variant were examined. We also studied the distribution of this hCNT1 intron retention transcript in several normal human tissues and some tumors, and observed variations in expression.
Materials and methods
Plasmid construction
RT-PCR was carried out with total RNA from human kidney (ClonTech) as a template using primers designated from the reported sequence of hCNT1a (GenBank accession number U62966): sense-EcoRV-hCNT1—5′-AGC GAT ATC TGG GAC ATG GAG AAC GAC-3′, antisense-SalI-hCNT1—5′-TTA GTC GAC TGT TCT GTC CTC ACT GTG CAC-3′ (restriction sites underlined). The PCR product was subcloned into FLAG tagged pCMV vector (Stratagene). Colony PCR was used to screen the positive colonies and a longer product was detected and sequenced (Molecular Resources Center, University of Tennessee Health Science Center, Memphis, TN).
Cell culture
Porcine kidney tubular epithelium nucleoside transporter deficient cells (PK15NTD) were kindly donated by Dr. Chung-Ming Tse (Johns Hopkins University, Baltimore, MD). Human pancreatic cancer Panc-1, HPAC II and MIA PaCa-2 cell lines were purchased from ATCC (Manassas, VA). Cells were maintained in Eagle’s minimal essential medium/Earle’s Balanced Salt Solution with 0.1 mM nonessential amino acids, 1 mM sodium pyruvate (PK15NTD), Dolbeco’s minimum essential medium (Panc-1 and MIA PaCa-2) and 10% fetal bovine serum (Invitrogen, Grand Island, NY) and 2.5% horse serum additionally for MIA PaCa-2), Dulbecco’s modified Eagle’s medium and Ham’s F12 medium containing 1.2 g/l sodium bicarbonate, 2.5 mM l-glutamine, 15 mM HEPES and 0.5 mM sodium pyruvate supplemented with 0.002 mg/ml insulin, 0.005 mg/ml transferrin, 40 ng/ml hydrocortisone, 10 ng/ml epidermal growth factor and 5% fetal bovine serum (HPACII) at 37 °C in a humidified atmosphere of a mixture of 5% CO2 and 95% air.
RNA extraction and RT-PCR
Total RNA of tumor cells (ATCC, Manassas, VA) was prepared using TRIzol reagent. 2 μg total RNA was treated with DNase I for 15 min to remove any genomic DNA that might be present, and subsequently used for cDNA synthesis. DNase-treated Human tissue total RNA was obtained from ClonTech, and cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen). PCR amplification was performed using the hCNT1 intron 4 primers. For tissue expression analysis, human tissue RNA arrays were purchased from Ambion for the above RT-PCR analyses.
The LightCycler system (Roche) was used for PCR under the following conditions: 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 10 s. The relative level of amplified RNA was normalized to mRNA expression of the housekeeping gene, glyceraldehyde-3-phosphate (GAPDH) or beta-actin (β-actin). Gene expression was evaluated by the ΔCT method using the GAPDH reference gene. All samples were amplified in duplicate and two non-template controls per primer pair were included in each run.
Tissues
Human normal and tumor tissue total RNAs, i.e. normal kidney, kidney tumor, normal liver, liver tumor, normal pancreas, were purchased from Ambion and used to analyze hCNT1 and hCNT1-IR transcript expression by real time RT-PCR.
Results
A novel splice variant of hCNT1 was identified in normal human kidney
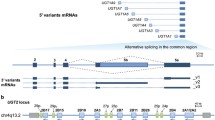
RT-PCR product of hCNT1 from normal human kidney total RNA obtained from a pool of 14 male/female Caucasians at ages between 18 and 58 years old was subcloned into a mammalian expression vector (pCMV-3flag). Colony PCR screening resulted in a longer product. Sequence alignment with full length hCNT1 cDNA showed that it contained an insertion of 734 bps from intron 4, between exons 3 and 4, and missing exons 5 and 13, as well as part of exon 6. We designated this novel splice variant of hCNT1 as hCNT1-IR (Fig. 1).
hCNT1-IR is not translated
To determine whether the hCNT1-IR mRNA encodes protein, we transfected PK15 nucleoside deficient cells [17] with the hCNT1-IR construct, similar to what we have described previously [18]. Western blot analysis using anti-flag antibody according to that procedure [18] showed no protein expression; which could be the reason for no uridine uptake in a uridine transport assay [18] (data not shown). This is not surprising as the introduction of the intron and missing of exons caused a frame shift that probably precludes stable protein translation. The sequence of the hCNT1-IR splice variant showing retention of intron 4 and skipping of some exons is shown in Fig. 2. A BLAST analysis of this transcript sequence showed several stop codons and the largest possible reading frame only showed half the length of the normal hCNT1 cDNA open reading frame, which would be a very severely truncated protein that would probably be unstable even if translated.
Sequence alignment of variant splice variant hCNT1-IR and normal hCNT1 transcripts. The top sequence is that of hCNT1-IR, while the bottom sequence corresponds to sequence of hCNT1. The initial and latter black bold face sequences are those of the initial and latter nucleotides of hCNT1-IR that are matched to corresponding to cDNA sequence of normal hCNT1 in black plain face nucleotides. In between these two regions and highlighted in yellow, are sequence of the retained intron 4 of hCNT1-IR in bold face purple, and the matching genomic sequence of normal hCNT1 in bold green sequence. Mutations in hCNT-IR sequence are indicated by red letters; insertions are indicated by blue letters, while deletions are indicated by cyan spaces. The skipped sequences are indicated by a blue dotted line
Tissue distribution and expression levels of hCNT1 intron 4 retention transcripts
The tissue expression pattern of this splice variant transcript was quantified by real-time PCR expression studies for its levels in 20 different human tissues, using GAPDH as internal standard. The PCR results as shown in Fig. 3, showed that the intron retention variant hCNT1-1R (Fig. 3b) was most abundant in small intestine, followed by kidney and liver, and the heart, but was not high in the other tissues. As compared with the normal hCNT1 variant expression (shown in Fig. 3a), the expression pattern of hCNT1-IR is similar, except the overexpression of the latter hCNT1-IR in heart tissue (Fig. 3b). In the original cloning of CNT1 from the rat [19], a high stringency multi-tissue northern blotting with rat tissues showed predominant expression in jejunum (small intestine) and kidney tissues but no significant expression in other tissues, but a later study by using immunoblotting showed expression of rCNT1 in liver tissue as well, albeit at lower level than in small intestine and kidney [20]. The distribution of this novel hCNT1 intron retention variant, hCNT1-IR also shares similarities with that of rCNT1 except like with hCNT1, its significant overexpression expression in heart tissues (Fig. 3). The meaning of this is currently not apparent but may be worth studying.
Tissue distribution of normal hCNT1-WT and intron retention variant transcript hCNT1-IR. For quantitative measurement of mRNA, DNase-treated human tissue total RNA human tissue total RNA panel was purchased from ClonTech. cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen). The PCR reactions using cDNA were performed in a LightCycler system (Roche) under the following conditions: 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 10 s. The relative level of amplified mRNA was normalized to mRNA expression of the housekeeping gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase). Gene expression was evaluated by the ΔCT method relative to the reference gene. All samples were amplified in duplicate and two non-template controls per primer pair were included in each run. a qRT-PCR analysis of different human tissue RNA that using primers for normal hCNT1 transcript. b qRT-PCR analysis of different human tissue RNA using primers for intron retention hCNT1-IR splice variant transcript. Data are the mean ± SEM for three experiments run in duplicates
Overexpression of hCNT1-IR splice variant in some tumor tissues and cancer cell lines
We also looked at the differential expression of the normal hCNT1 and of hCNT1-IR transcripts. For the most part, the expression patterns of the two transcripts were similar, with small intestine expressing the highest levels, and substantial expression in the kidney and liver. However, in these normal tissues, there was some significant differences in expression also, being that while the kidney expressed more hCNT1 than the lever (Fig. 3a), the two tissues expressed the same level of hCNT1-IR (Fig. 3b). It is also notable that while hCNT1 expression level was substantial in the placenta, hCNT1-IR expression was not. Also, hCNT1-IR was shown to be expressed in heart tissue, while hCNT1 was not (Fig. 3). The significance of the similarities and differences in tissue expression patterns of the hCNT1 and hCNT1-IR variants is not apparent at this time. Important also is the distribution among the few tumor tissues that were examined. As shown in Fig. 4 interestingly, while normal tissues expressed higher levels of hCNT1 normal variant than tumor tissues, the opposite was true for the hCNT1-IR splice variant, which was overexpressed in tumor tissues relative to normal ones. In a study by Pennykooke et al. with human tumor tissues [21], hCNT1 was overexpressed in some kidney tumors, but there was generally a decrease in relative expression compared to normal kidney tissue. Our RT-PCR results of the expression of the intron retention variant in tumor samples shows an overexpression of hCNT1-IR in human kidney and liver tumor tissues, as well as significant overexpression in human hepatocellular carcinoma (HepG2) cell line compared to normal kidney and liver tissues (Fig. 4 a, b). The overexpression of hCNT1-IR in cancer relative to normal tissues was also demonstrated with pancreatic cancer cells whereby there was a significant downregulation of hCNT in cancer cells (Fig. 4a), but not in hCNT1-IR (Fig. 4b). A comprehensive analysis of hCNT1-IR expression in tumors and matched normal tissue is warranted.
Comparative quantitative RNA analysis of hCNT1-IR or normal hCNT1 (hCNT-WT) transcript expression in human normal and tumor tissues or cancer cell lines. RNA was quantified from different tissues and cells using quantitative reverse transcription real-time-PCR (RT-qPCR). a Expression of normal hCNT1 transcript in normal human liver or normal human pancreas tissue (red), and in the human liver cancer cell line HepG2 or human pancreatic cancer cell lines Panc-1, HPAF-II and MIAPaCa2 (blue). b Relative expression of novel splice variant hCNT1-IR in the normal human liver and pancreas tissues (red) and in the human liver cancer cell line HepG2 and pancreatic cancer cell lines Panc-1, HPAF-II and MIAPaCa2 (blue). c Relative expression of novel splice variant hCNT1-IR in normal human liver or kidney tissue (red) and commercially available liver or kidney tumor tissue (blue). Each data point represents the mean and standard deviation of the mean from two independent experiments run in triplicate samples. *p < 0.05, **p < 0.01
Discussion
The hCNT1 transporter protein is a widely expressed, high-affinity, pyrimidine-preferring, nucleoside transporter implicated in the uptake of physiological pyrimidine nucleosides as well as a variety of derivatives used in anticancer and antiviral treatment. hCNT1 is generally downregulated in cancer tissues and suspected of tumor suppressor activity. Here, we report a novel splice variant of hCNT1, hCNT1-intron retaining designated hCNT1-IR in human kidney and cancers. In hCNT1-IR, a 734-base pair sequence in the fourth intron of the hCNT1 gene was spliced between exons 3 and 4, while exons 5, 13 and part of exon 6 were skipped. hCNT1-IR protein was not detected in cells, and consequently no functional activity was observed. Interestingly, this intron-retention, exon skipping variant of hCNT1 was overexpressed not only in kidney but also in liver tumor, hepatocellular carcinoma and pancreatic cancer cells. This novel splice variant may be associated with pathological conditions of cancer, particularly kidney, liver and pancreas cancers, and cell lines, including HepG2, Panc-1, HPAC-II and MiaPaCa2 cells. Although the biological function of this variant remains to be elucidated, this is the first report of exon skipping and intron retention in nucleoside transporter RNA aberrant splicing. This study suggests that overexpression of this hCNT1 splice transcript may serve as a cancer biomarker.
Alternative splicing allows the production of multiple mRNA forms from one gene and therefore is crucial for protein diversity [22]. In addition, alternative splicing contributes to tissue and developmental stage-specific expression of protein isoforms. Besides yielding different proteins, alternative splicing in the 5′-noncoding region will not affect the gene product, but may influence gene expression and translation [22, 23]. Increasing evidence suggests that alterations in RNA processing can lead to a variety of human diseases, including cancer [22, 24]. It has been estimated that at least 15% of mutations that cause genetic diseases affect pre-mRNA splicing [25]. Indeed, numerous studies have indicated that alternative splicing occurs frequently in cancer cells, and a plethora of cancer-specific splice variants have been reported [26,27,28]; and some of them are considered to be candidate cancer biomarkers, such as CD44 [28] and the Wilms tumor (WT1) genes [29]. The new hCNT1 splice variant did not show expression of protein, suggesting that the retained intron might restrict its translation or lead to frame shifts and/or stop codons that might prevent translation of stable protein(s). In this regard it has been reported that a frameshift deletion in the human equilibrative nucleoside transporter 3 (hENT3) coded by the SLC29A3 gene led to a paradoxical translation of an otherwise noncoding mRNA splice variant [30]. The tissue expression pattern of the aberrant splice variant, hCNT1-IR is similar to that of the normal hCNT1. Intriguingly though, the aberrant splice variant is overexpressed in tumor tissue relative to normal tissues of the same organ origin as demonstrated by our analysis of normal and tumor tissues of kidney and liver, in particular. This is opposite to the general downregulation of hCNT1 in cancers. This suggests that the aberrant hCNT1 splicing may be overactive in disease conditions; at least in cancer. However, its physiological significance and possibly pathological function still need to be studied. Especially, it will be important to study the significance of hCNT1 aberrant splicing in the pathogenesis of tumors that express it, like kidney and liver cancers. Thus, as we propose, the hCNT1-IR splice variant might serve as a novel tumor biomarker, and possibly a therapeutic target. In broader terms, this study might open the door to studying such complex splicing of nucleoside transporter (NT) genes. Our study shows for the first time the combined intron retention and exon skipping splicing of any nucleoside transporter (NT) gene. A recent report described an insertion mutant of the hCNT3 (SLC28A3) gene that incorporated an extra 176 base pair nucleotide segment within exons 2 and 3 with the expressed a protein confined to the endoplasmic reticulum [31]. However, to date no intron retention and/or exon skipping nucleoside transporter transcript has been reported; and no variations in non-coding regions that might alter mRNA stability or transcription efficiency through polymorphisms in intron, or promoter regions or splice variation of NT genes have been described.
It is important to note that in sporadic amyotrophic lateral sclerosis (ALS) patients, multiple abnormal glutamate transporter EAAT2 (GLT-1) variant splicing with intron retention or exon skipping has been shown to result in the loss of EAAT2 protein activity but contributes to pathogenesis of the disease [32]. It is thus possible that in a similar manner, nucleoside transporter splicing such as we have unraveled in our study may contribute to pathogenesis of diseases linked to nucleoside homeostasis.
Our current study potentially opens the door to studying aberrant splicing in NT gene regulation, and in pathogenesis. It has been demonstrated that splice variant expression can be used as a means to regulate gene expression. An example is the EAAT2 transporter mentioned above, whereby abnormal splicing of the EAAT2 RNA repressed the normal EAAT2 protein expression [33]. This suppression of EAAT2 expression occurred to protect undifferentiated glioma cells from death due to the uptake of glutamate [32]. The rat, CNT1 gene expression is downregulated in liver tumors relative to normal tissue, with CNT1 protein being even absent in tumor lesions [34]. It has also been reported that hCNT1 is downregulated in human breast [35] and gynecological [36] cancers. In the gynecological cancer study, hCNT1 was by far the hCNT subtype that was most frequently downregulated or lost in the tumors analyzed. Moreover, hCNT1 protein loss was highly correlated with poor disease prognosis [35]. Conversely, the overexpression of hCNT1 has been shown to inhibit pancreatic cancer cell growth and increase sensitivity to chemotherapy [37]. The mechanism of downregulation of the usual hCNT1 variant in cancers noted above is unknown. As the aberrant splice variant expression of the EAAT2 glutamate transporter is used to suppress the normal variant expression in glioma cells [33, 38], the aberrant splice variant expression of hCNT1-IR may also be used to downregulate the expression on the normal hCNT1 variant in cancer.
Conclusions
In conclusion, we have for the first time identified a novel splice variant transcript of hCNT1 that retains intron 4, skips exons 5 and 6, and part of exon 13. The tissue distribution of this new splice variant (hCNT1-IR) is similar to normal hCNT1 except in a few tissues like placenta which expresses hCNT1 and not hCNT1-IR, and heart tissue, which expresses hCNT1-IR but not normal hCNT1. Importantly, opposite to the pattern of normal hCNT1, which is usually downregulated in cancers, hCNT1-IR is overexpressed in some cancers relative to normal tissues. This could lay the foundation for developing a new cancer biomarker and/or cancer therapeutic target.
Availability of data and materials
The datasets collected and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CNTs:
-
concentrative nucleoside transporters
- ENTs:
-
equilibrative nucleoside transporters
- hCNT1:
-
human concentrative nucleoside transporter-1
- hCNT1-IR:
-
human concentrative nucleoside transporter-1 intron retention
- NT:
-
nucleoside transporter
- SLC28A1 :
-
solute carrier gene family 28 variant A1
- SNPs:
-
single nucleotide polymorphisms
References
Cheeseman CI, Mackey JR, Cas CE, Baldwin SA, Young JD. Molecular mechanisms of nucleoside and nucleoside drug transport, in a volume of current topics in membranes. San Diego: Academic Press; 2000. p. 330–79.
Mackey JR, Baldwin SA, Young JD, Cass CE. Nucleoside transport and its significance for anticancer drug resistance. Drug Resist Updates. 1998;1:310–24.
Ritzel MW, Yao SY, Huang MY, Elliott JF, Cass CE, Young JD. Molecular cloning and functional expression of cDNAs encoding a human Na+ nucleoside cotransporter (hCNT1). Am J Physiol. 1997;272:C707–14.
Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 2004;447:728–34.
Graham KA, Leithoff J, Coe IR, Mowles D, Mackey JR, Young JD, Cass CE. Differential transport of cytosine-containing nucleosides by recombinant human concentrative nucleoside transporter protein hCNT1. Nucleosides Nucleotides Nucleic Acids. 2000;19:415–34.
Lostao MP, Mata JF, Larrayoz IM, Inzillo SM, Casado FJ, Pastor-Anglada M. Electrogenic uptake of nucleosides and nucleoside-derived drugs by the human nucleoside transporter 1 (hCNT1) expressed in Xenopus laevis oocytes. FEBS Lett. 2000;481:137–40.
Yao SY, Ng AM, Sundaram M, Cass CE, Baldwin SA, Young JD. Transport of antiviral 30-deoxy-nucleoside drugs by recombinant human and rat equilibrative, nitrobenzylthioinosine (NBMPR)-insensitive (ENT2) nucleoside transporter proteins produced in Xenopus oocytes. Mol Membr Biol. 2001;18:161–7.
Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, Kroetz DL, Ferrin TE, Clark AG, Risch N, Herskowitz I, Giacomini KM. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci. 2003;100:5896–6901.
Nozawa T, Nakajima M, Tamai I, Noda K, Nezu J, Sai Y, Tsuji A, Yokoi T. Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J Pharmacol Exp Ther. 2002;302:804–13.
Mata JF, Garcia-Manteiga JM, Lostao MP, Fernandez-Veledo S, Guillen-Gomez E, Larrayoz IM, Lloberas J, Casado FJ, Pastor-Anglada M. Role of the human concentrative nucleoside transporter (hCNT1) in the cytotoxic action of 5-deoxy-5-fluorouridine, an active intermediate metabolite of capecitabine, a novel oral anticancer drug. Mol Pharmacol. 2001;59:1542–8.
Gray JH, Mangravite LM, Owen RP, Urban TJ, Chan W, Carlson EJ, Huang CC, Kawamoto M, Johns SJ, Stryke D, et al. Functional and genetic diversity in the concentrative nucleoside transporter, CNT1, in human populations. Mol Pharmacol. 2004;65:512–9.
Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–65.
Roy SW, Irimia M. Splicing in the eukaryotic ancestor: form, function and dysfunction. Trends Ecol Evol. 2009;24:447–55.
Clark F, Thanaraj TA. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet. 2002;11:451–64.
Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–45.
Maekawaa S, Moria D, Nishiyaa T, Takikawab O, Horinouchia T, Nishimotoa A, Kajitaa E, Miwaa S. OCTN2VT, a splice variant of OCTN2, does not transport carnitine because of the retention in the endoplasmic reticulum caused by insertion of 24 amino acids in the first extracellular loop of OCTN2. Biochim Biophys Acta. 2007;1773:1000–6.
Ward JL, Sherali A, Mo ZP, Tse CM. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J Biol Chem. 2000;275:8375–81.
Wang C, Pimple SR, Buolamwini JK. Interaction of benzopyranone derivatives and related compounds with human concentrative nucleoside transporters 1, 2 and 3 heterologously expressed in porcine PK15 nucleoside transporter deficient cells. Structure–activity relationships and determinants of transporter affinity and selectivity. Biochem Pharmacol. 2010;79:307–20.
Huang QQ, Yao SY, Ritzel MW, Paterson AR, Cass CE, Young JD. Cloning and functional expression of a complementary DNA encoding a mammalian nucleoside transport protein. J Biol Chem. 1994;269:17757–60.
Hamilton SR, Yao SY, Ingram JC, Hadden DA, Ritzel MW, Gallagher MP, Henderson PJ, Cass CE, Young JD, Baldwin SA. Subcellular distribution and membrane topology of the mammalian concentrative Na+-nucleoside cotransporter rCNT1. J Biol Chem. 2001;276:27981–8.
Pennycooke M, Chaudary N, Shuralyova I, Zhang Y, Coe IR. Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem Biophys Res Commun. 2001;280:951–9.
Lou H, Gagel RF. Alternative ribonucleic acid processing in endocrine systems. Endocr Rev. 2001;22:205–25.
Kozak M. Do the 5′untranslated domains of human cDNAs challenge the rules for initiation of translation (or is it vice versa)? Genomics. 2000;70:396–406.
Kawakubo K, Kuhen KL, Vessey JW, George CX, Samuel CE. Alternative splice variants of the human PKR protein kinase possessing different 5′-untranslated regions: expression in untreated and interferon-treated cells and translational activity. Virology. 1999;264:106–14.
Shalev A, Blair PJ, Hoffmann SC, Hirshberg B, Peculis BA, Harlan DM. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002;143:2541–7.
Philips AV, Cooper TA. RNA processing and human disease. Cell Mol Life Sci. 2000;57:235–49.
Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54.
Naor D, Nedvetzki S, Golan I, Melnik L, Faitelson Y. CD44 in cancer. Crit Rev Clin Lab Sci. 2002;39:527–79.
Wagner KD, Wagner N, Schedl A. The complex life of WT1. J Cell Sci. 2003;116:1653–8.
Bolze A, Abhyankar A, Grant AV, Patel B, Yadav R, Byun M, Caillez D, Emile JF, Pastor-Anglada M, Abel L, Puel A, Govindarajan R, de Pontual L, Casanova JL. A mild form of SLC29A3 disorder: a frameshift deletion leads to the paradoxical translation of an otherwise noncoding mRNA splice variant. PLoS ONE. 2012;7:e29708.
Errasti-Murugarren E, Molina-Arcas M, Casado FJ, Pastor-Anglada M. A splice variant of the SLC28A3 gene encodes a novel human concentrative nucleoside transporter-3 (hCNT3) protein localized in the endoplasmic reticulum. FASEB J. 2009;23:172–82.
Lin CG, Bristol LA, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein JD. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602.
Guo H, Lai L, Butchbach MER, Lin C-LG. Human glioma cells and undifferentiated primary astrocytes that express aberrant EAAT2 mRNA inhibit normal EAAT2 protein expression and prevent cell death. Mol Cell Neurosci. 2002;21:546–56.
Dragan Y, Valdés R, Gomez-Angelats M, Felipe A, Javier Casado F, Pitot H, Pastor-Anglada M. Selective loss of nucleoside carrier expression in rat hepatocarcinomas. Hepatology. 2000;32:239–46.
Lane J, Martin TA, McGuigan C, Mason MD, Jiang WG. The differential expression of hCNT1 and hENT1 in breast cancer and the possible impact on breast cancer therapy. J Exp Ther Oncol. 2010;8:203–10.
Farré X, Guillén-Gómez E, Sánchez L, Hardisson D, Plaza Y, Lloberas J, Casado FJ, Palacios J, Pastor-Anglada M. Expression of the nucleoside-derived drug transporters hCNT1, hENT1 and hENT2 in gynecologic tumors. Int J Cancer. 2004;112:959–66.
Bhutia YD, Hung SW, Patel B, Lovin D, Govindarajan R. CNT1 expression influences proliferation and chemosensitivity in drug-resistant pancreatic cancer cells. Cancer Res. 2011;71:1825–35.
Ye ZC, Rothstein JD, Sontheimer H. Compromised glutamate transport in human glioma cells: reduction-mislocalization of sodium-dependent glutamate transporters and enhanced activity of cystine–glutamate exchange. J Neurosci. 1999;19:10767–77.
Acknowledgements
We like to thank the University of Tennessee, the Rosalind Franklin University of Medicine and Science for facilities support and the National Institutes of Health (NIH) for funding support.
Funding
NIH Grant Nos. CA-125850 and R01-GM104503.
Author information
Authors and Affiliations
Contributions
JKB and CW designed the experiments. CW did the experiments and collected the data. CW and JKB analyzed the data. CW drafted the manuscript and JKB edited it. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable because the human tissues were purchased from commercial sources with no identification.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, C., Buolamwini, J.K. A novel RNA variant of human concentrative nucleoside transporter 1 (hCNT1) that is a potential cancer biomarker. Exp Hematol Oncol 8, 18 (2019). https://doi.org/10.1186/s40164-019-0144-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40164-019-0144-y