Abstract
Spermatogenesis is a complex differentiating developmental process in which undifferentiated spermatogonial germ cells differentiate into spermatocytes, spermatids, and finally, to mature spermatozoa. This multistage developmental process of spermatogenesis involves the expression of many male germ cell-specific long noncoding RNAs (lncRNAs) and highly regulated and specific gene expression. LncRNAs are a recently discovered large class of noncoding cellular transcripts that are still relatively unexplored. Only a few of them have post-meiotic; however, lncRNAs are involved in many cellular biological processes. The expression of lncRNAs is biologically relevant in the highly dynamic and complex program of spermatogenesis and has become a research focus in recent genome studies. This review considers the important roles and novel regulatory functions whereby lncRNAs modulate mammalian spermatogenesis.
Similar content being viewed by others
Introduction
Mammalian spermatogenesis is the process by which male germline cells divide and differentiate into mature spermatozoa [1]. It is an androgen-dependent process that overrides somatic cell and germ cell interactions [2]. Spermatogenesis is a complex physiological process involving the division, differentiation, and meiosis of immature male germ cells into mature haploid spermatozoa in the seminiferous tubules of the testis [3]. This process includes acrosomal formation, elimination of cytoplasm, chromatin reorganization, and flagellum development, replacement of protamines, removal of histone from chromatin material and nuclear formation [2, 3]. Spermatogenesis is also closely related to the Sertoli cells of the seminiferous tubules. Sertoli cells and germ cells produce doublesex and Mab-3 related transcription factor 1 (DMRT1), which is an evolutionary conserved transcriptional factor that regulates tubule morphology and other spermatogonial functions through its Sertoli cell maturation and polarity effects [4]. Lei et al. reported a decreasing trend in the number of cells in seminiferous tubules after silencing lncRNA H19 (H19 imprinted maternally expressed transcript) [5]. This affected the expression of insulin-like growth factor receptor (IGF-1R) in Sertoli cells and Spermatogenic cells. IGF-1 maintains the survival of several types of stem cells and has essential functions in male reproduction [5]. After the completion of germ cell development into elongated spermatids, the mature spermatids move from the Sertoli cells into the tubule lumen through rete testis until they reach the efferent ducts of the epididymis. The round nucleus of the developing spermatid starts to elongate and condense to achieve a spindle shape during the morphogenesis stage and spermatid develop into long needle-shaped mature sperm [6]. In this complicated developmental process, the mature spermatids execute complex biochemical activities to reach the stage of motile spermatozoa. The dynamic process of spermatogenesis is highly regulated by tissue and cell-specific gene expression. During this precisely regulated biological process, phase-specific gene expression is controlled post-transcriptionally by long noncoding RNAs (lncRNAs) [1, 7]; however, little is known about the regulation of spermatogenesis by lncRNAs in humans [8].
Mammals transcribe a large amount of RNAs; however, only a small fraction (around 2%) of transcripts encode proteins. The remaining large number of transcripts represent small noncoding RNAs and lncRNAs [7, 9]. LncRNAs are a recently discovered class of non-coding transcripts [6], ranging in size from 200 bp to 100,000 bp, and most of them exhibit cell-type-specific expression [10,11,12]. LncRNAs lack an open reading frame in some or all of their sequence and have no protein-coding ability [7]. They are transcribed in the nucleus and cytoplasm by RNA polymerase II and are spliced, capped, and polyadenylated [13,14,15].
The molecular functions of lncRNAs have been studied in different cell lines [16, 17] by lncRNA knockdown and overexpression, and include regulation of protein activities, organizational roles, serving as precursors for small RNAs, and modulating transcriptional patterns [18]. LncRNAs are involved in the regulation of many complex cellular and molecular processes during development [6], gene imprinting, and X chromosome inactivation [16]. The majority of lncRNAs are expressed at low levels and generally exhibit low primary sequence conservation. Few studies have demonstrated genetic evidence that mammalian lncRNAs function in in vivo of animal models [6, 19]. LncRNAs often demonstrate restricted time-specific or tissue-specific developmental expression patterns at lower levels than mRNAs. LncRNAs exert their various regulatory functions and biological processes by interacting with splicing factors and recruiting transcription factors. During this process, lncRNAs may alter mRNA splicing and affect gene expression. They may also be involved in transcriptional activation, transcriptional repression, epigenetic modulation, and RNA splicing regulation.
Many lncRNAs have been noted as being involved in spermatogenesis; however, very few of them have been validated and functionally characterized. Thus, the majority of lncRNAs expressed during mammalian spermatogenesis need to be validated and their molecular functions should be specified experimentally. This paper discusses the expression patterns and known functions of certain lncRNAs and their suggested roles in mammalian spermatogenesis. The main purpose of this review was to provide a better understanding of mammalian spermatogenesis in terms of lncRNAs and to explain the classification, expression, and potential roles of lncRNAs in regulating spermatogenesis.
The developmental processes of mammalian spermatogenesis
The developmental process of mammalian spermatogenesis involves the continuous production of functional sperm, starting from spermatogonial stem cells and involving different cell types. Spermatogenesis involves a series of differentiation of morphologically undifferentiated tissues and cells into highly functional cells, including self-renewing stem cells [2]. The expression of genes and non-coding RNAs during spermatogenesis leads to the production of proteins that maintain the overall housekeeping functions and perform specific processes in the germ cell developmental stages, respectively [20, 21].
There are three principal phases or stages of spermatogenesis: Mitosis, meiosis, and post meiosis stages. Spermatogonial cells undergo mitosis for self-renewal and proliferation, which promotes spermatogonial differentiation. During the second stage, meiosis and genetic recombination occur in the spermatocytes. No further replication occurs and the haploid male germ cells differentiate in the post-replicative stage into spermatids. In mice, the mitotic, meiotic, and post meiotic phases last around 10, 11, and 14 days, respectively. The spermatogenic stem cells differentiate six times during the mitotic phase to form type A spermatogonia, intermediate spermatogonia, and type B spermatogonia. The final division of this phase produces preleptotene spermatocytes that commence meiotic division and undergo the S phase of the spermatogenesis in their last cell cycle. The second meiotic stage is succeeded by two meiotic divisions producing spermatids in the post-meiotic stage. At this stage, DNA replication does not occur and the spermatids develop into fully mature spermatozoa via spermiogenesis by remodeling through the process of acrosome formation, condensation of the nucleus, development of flagellum, and loss of a large portion of the cytoplasm [22,23,24].
Thus, spermatogonial stem cells undergo mitotic divisions giving rise to meiotic spermatocytes. Then, the reductive divisions of meiosis lead to the formation of round spermatids (haploid). The round spermatids undergo a series of differentiation to develop into elongated spermatids and then fully develop into mature spermatozoa (Fig. 1).
Classification of lncRNA
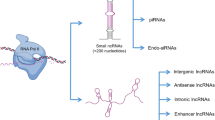
Post-genomic studies have identified a diversity of transcriptional products, such as many small regulatory RNAs and a large number of polyadenylated and non-polyadenylated lncRNAs. These lncRNAs are classified according to their protein-coding loci as intergenic, antisense, intronic, or overlapping lncRNAs. Novel lncRNAs may be similarly classified and subtyped as intergenic, ambiguous, antisense, and intronic lncRNAs according to their genomic locations [22] as shown in Fig. 2. Mattich and Rinn also reviewed the classification of lncRNAs by the relative location of their transcripts according to the adjacent protein-coding genes as intergenic, intronic, bidirectional, overlapping antisense, and overlapping sense lncRNAs [13]. Some lncRNAs are transcribed within long terminal repeats (LTR). Many other lncRNAs do not originate from bidirectional mRNA promoters and may have promoters in their intergenic regions that do not overlap with the LTRs and lncRNA promoters [23].
Structural classification of lncRNAs
LncRNAs can be categorized into three groups according to their relationship with the adjacent protein-coding genes [24,25,26] as indicated in Fig. 3.
1) Antisense or sense: These lncRNAs are located and transcribed on the opposite or the same strand of the adjacent protein-coding genes [27]. 2) Convergent (divergent): These lncRNAs have a convergent (divergent) orientation of transcription compared with that of the adjacent protein-coding genes [26]. 3) Intergenic (intronic): These lncRNAs are located between two protein-coding genes, or reside in introns [26, 28].
Functional classification of lncRNAs
Most lncRNAs are non-coding, but play diverse roles in many biological processes and are associated with disease occurrence [29]. LncRNAs may function through DNA, RNA, and protein interactions; however, the precise molecular functions of most lncRNAs are unknown. Unlike protein-coding genes, the lncRNA sequence does not have sequence motifs that indicating their function, and their secondary structures are not conserved [25]. This makes it more difficult to predict the function of lncRNAs based on their sequence motifs, conserved sequences, and secondary structures. Current studies indicate that the function of lncRNAs in cell-based studies can through their regulation of, and interactions with, protein-coding genes, microRNAs (miRNAs), and other lncRNAs, using different approaches (Table 1).
The lncRNAs use different mechanisms to carry out complex functions and have played some of the following roles [27] as shown in Fig. 4.
-
I)
Signaling: The transcriptional activity of lncRNAs can be described by their response to diverse stimuli, the production of signals, and cell type-specific expression. The molecular mechanism of the majority of the lncRNAs may indicate considerable transcriptional control on a molecular basis by RNA polymerase II. The occurrence of lncRNAs during the developmental processes of specific tissues and cells at a specified time demonstrates the effectiveness of transcriptional control by interpreting the cellular context as well as gene regulation and acts as a molecular signal [14]. The lncRNA-associated signaling pathways indicate the role of transcription factors in gene regulation at a specific time and space for the signaling archetype lncRNAs. De Santa et al. used chromatin immunoprecipitation sequencing (ChIP-seq) to show that lncRNA transcripts can be produced by gene activating enhancers and that their expression correlated positively with the expression level of their neighboring genes, indicating mRNA synthesis regulation [32] as indicated in Fig. 4.
-
II)
Guides: The interaction of lncRNAs with their target molecules may enhance the proper positioning as the transcription machinery on adjacent genes by guiding, in cis and in trans, for distantly located genes [14]. A lncRNA, as a guide molecule, attaches to the RNA binding complex to guide the complex and regulates the expression of the gene at its genomic locus (see Fig. 4).
-
III)
Decoys: Enhancers and promoters play an important role in transcriptional regulation of lncRNAs, both negatively and positively [33]. Effectors can be negatively regulated by lncRNAs, which function as molecular decoys (see Fig. 4). The telomeric repeat-containing RNA (TERRA), which is large noncoding RNA, interacts with the telomerase protein to inhibit its function [34] and the release of the receptor from the DNA of the growth arrest-specific 5 (Gas5) gene via a hairpin sequence motif that represses the glucocorticoid receptor [35], are examples of lncRNAs functioning as a molecular decoys. In addition, the proliferation of spermatogonial stem cells is promoted by lncRNA-AK015322 by acting as decoy for microRNA-19b-3p [36].
-
IV)
Scaffold: The lncRNAs form a distinct complex from two or more proteins to act as a molecular scaffold as shown in Fig. 4. The scaffold complexes may play an important function in controlling and enhancing cellular signals and intermolecular interactions [31]. The scaffold type lncRNAs bind with different domains of the effector components to form a nucleoprotein complex (see Fig. 4). These complexes may be involved in gene activation [28], chromatin modification, and gene repression [37].
Databases of lncRNAs
Comprehensive datasets play an important role in facilitating the classification, validation, functional prediction, and assessment in low and high throughput lncRNA experiments. Evidence for the biological properties of well-studied and identified lncRNAs has been deposited in specialized databases that have been developed to provide and improve information resources for lncRNA research in the public domain. These developed lncRNA databases have been designed to contain important and comprehensive information about interactions among macromolecules, genomic structure, conservation potential, expression profile, epigenetic modifications, and functional annotations [38].
The lncRNA databases can be categorized as annotation, interaction, and specific databases as shown in Table 2. The annotation databases include NONCODE v4.0, lncRNAdb, LNCipedia, lncRNome, fRNAdb, lncRNAtor, lncRNAMap, and PLncDB. The ChIPBase, NPInter, miRcode, DIANA-LncBase, StarBasev2.0, lncRNA2Target, and lncRNADisease are examples of interaction databases. The specific databases include lnCeDB, NRED, Linc2go, and lncRNASNP [49,50,51,52,53,54,55,56,57,58,59].
Identification of lncRNAs during spermatogenesis
LncRNAs can be identified and functionally characterized systematically in different ways [6]. Genome-wide transcriptome analyses [60, 61] RNA-Seq approach [62], RNA capture sequencing [63], and the development of efficient gene knock out technologies like the Clustered regularly interspaced short palindromic/Cas9: CRISPR-associated protein-9 nuclease (CRISPR/Cas9) with a homologous recombination system [6] could expand our understanding of lncRNAs’ biological functions in spermatogenesis. The optimized CRISPR system plays an important role in targeting gene replacement and knock out up to 92 kb to study gene function and is successful because of its low rate of off-target effects. Phylogenetic, conservational, and comparative genomic analyses will also help to determine the evolution and origin of testis enriched lncRNAs. Cell-based in vitro studies of genomic loss of function [64], including targeted in vivo lncRNA silencing and deletion revealed the mechanisms of lncRNAs in cellular processes and development [6, 65, 66]. See the descriptions of the other identification methods of lncRNA in Table 3.
The expression of lncRNAs during mammalian spermatogenesis
Mammalian spermatogenesis plays an important role in fertility and the continual replacement of species from generation to generation. Previously, many studies have analyzed the molecular mechanisms of mammalian spermatogenesis, focusing on microRNAs, Piwi-interacting RNAs (piRNAs), and protein-coding genes; however, limited studies have been performed on lncRNAs in mammalian spermatogenesis. Some studies reported testis and germ cells expression profiles of lncRNAs in different ages and stages of development [52]. Bao et al. analyzed lncRNAs in fetal and postnatal mouse testes using microarray-based profiling to investigate the novel links between lncRNAs and their targets [52]. The results suggested the involvement of lncRNAs in gene regulation at transcriptional and post-transcriptional levels. Sun et al. conducted a microarray analysis to assess the lncRNA expression profiles during postnatal development in mouse testis [21]. Song et al. explored the testis-specific lncRNAs and their expression in adult mouse testis using experimental and computational methods [53] and reported highly expressed lncRNAs in adult testis. Wichman et al. analyzed the dynamic expression pattern of lncRNAs in the testis that escapes the meiotic sex chromosome inactivation (MSCI) [54]. The X- and Y- linked lncRNAs showed higher expression in pachytene spermatocytes, suggesting their involvement in escape from MSCI. In that study, RNA-Seq analysis confirmed that a small number of mRNAs showed higher expression patterns in the pachytene stage, indicating that few mRNAs escape MSCI.
Recent studies have assessed the lncRNA expression during spermatogenesis. Many lncRNAs show different expression levels and tissue specificities in different stages of mammalian spermatogenesis [55], suggesting that they have various functions in different biological processes, such as in the regulation of gene silencing, cell division, gonadogenesis, and sex determination. Zhang et al. reported the expression of lncRNAs in human normal sperm and in asthenozoospermic patients. The results suggested that enriched lncRNAs have an important role in sperm motility [56]. Some of the lncRNAs, such as lnc32058, lnc09522, and lnc98487 are upregulated in asthenozoospermic sperm, exhibiting specific expression patterns in sperm and testis and are related to the progressive motility of sperm, providing insights into the causes of male infertility [56]. Zhang et al. revealed that the low expression of lncRNA HOTAIR (Hox transcript antisense intergenic RNA) in the spermatozoa of patients with asthenozoospermia and oligoasthenozoospermia, which might be related with the motility and vitality of sperm [57]. Evolutionary conservation analysis of hsa-lncRNA12238 demonstrated its highest expression in the human testis, indicating its involvement in spermatogenesis, sperm-egg recognition, and other reproductive processes [58].
Liang et al. analyzed the overall sequential expression of lncRNAs in four specific stages of spermatogenesis, mainly in spermatogonial stem cells (SSC), type A spermatogonia (AS), pachytene spermatocytes (PS), and round spermatids (RS) in mice using microarray analysis. They reported that intergenic lncRNAs were highly expressed in the germ cell types, acting as tissue-specific lncRNA genes and as housekeeping genes, sense overlap lncRNA, antisense lncRNA, and bidirectional lncRNA [59]. Furthermore, correlation analysis between mRNAs and lncRNAs showed high correlation coefficients and suggested coordinated changes in their expression during the biogenesis and function of male spermatogenesis. A similar investigation reported the expression of antisense lncRNA 1700108J01Rik and long intergenic noncoding RNA 1700101O22Rik [53] in meiotic prophase and round spermatid stages in testicular germ cells. Similarly, Dai et al. reported higher expression of lncRNA-testicular cell adhesion molecule 1 (lncRNA-Tcam1) in round spermatids than in somatic cells or germ cells in mice, with lncRNA-Tcam1 being localized in 45% of the cells found in testis [55]. See the expressions of some lncRNAs in Table 4.
Therefore, increasing amounts of research have demonstrated the dynamic changes in gene expression in the complex process of spermatogenesis. The findings of these studies provide new insights into lncRNAs expression levels and their tissue specificity in testicular spermatogenesis, suggesting their specific roles in mammalian spermatogenesis.
Functional roles of lncRNAs during mammalian spermatogenesis
LncRNAs are mRNA-like transcripts of greater than 200 nucleotide sequences exhibiting little or no protein-coding ability [83] and represent poorly understood ncRNAs. However, lncRNAs and protein-coding genes exhibit conserved co-expression networks during the division and differentiation of male germline stem cells into mature sperm [84]. The testis-specific lncRNAs evolved faster and showed higher conservation of post-transcriptional sequences during germ cell differentiation. The lncRNAs regulated genes are mostly associated with metabolic and reproductive activities, indicating the influence of lncRNAs on the control of gene expression during germ cell development. The transcriptional regulatory functions of testis-specific lncRNAs were predicted to control the expression of genes in proximal and distal positions [6].
Wen et al. reported the partial and full rescue of testis-specific lncRNAs in a trans-configuration using transgenes, indicating their DNA regulatory functions in trans during late spermatogenesis [6]. This study also indicated the functional role of certain testis-specific lncRNAs in the control of nuclear condensation efficiency, shaping, and spermatid differentiation or individualization. Other lncRNAs are via an in cis-mediated mechanism to regulate neighboring gene expression [85, 86]. Song et al. reported the cytoplasmic distribution of testis-specific lncRNAs involved in post-transcriptional gene regulation [53].
Mammals exhibit condensation and remodeling of their chromatin material during late spermatogenesis by omitting excess cytoplasm and replacing histones with protamine for spermatid individualization, leading to a highly compact sperm nucleus [64]. Enhancer-associated lncRNAs participate in transcriptional activation by acting over long distances on distal promoters associated with protein factors and the modulation of chromatin structures [87]. Song et al. revealed the cytoplasmic distribution of 1700108J01Rik and 1700101O22Rik using in situ hybridization, suggesting their function in post-transcriptional gene regulation mainly in the nucleus; however, they are not involved in epigenetic and transcriptional regulation [53]. Similarly, Zhang et al. reported the post-transcriptional role of lncRNA Dmrt1 on chromosome 5 in spermatogenesis and testis development in mice [65]. A study by Liang et al. revealed that the identified specific lncRNAs in spermatogonial stem cells play important roles in maintaining these cells, and some of them are involved in the regulation of differentiation of specific stages of testicular spermatogenesis and germ cell meiosis [80]. The authors concluded that the transcription mechanism of the mRNA and lncRNA play a key role in the differentiation and meiosis of mouse male germ cells. This study also provided comprehensive data on the coordinated changes in total lncRNA/mRNA transcription regulation, suggesting their importance in reproductive disorder diagnosis and treatment. The role of some lncRNAs in mammalian spermatogenesis is shown in Table 5.
Molecular signaling pathways enriched in lncRNA modulated spermatogenesis
LncRNAs play an important role in diverse biological processes by interacting with regulatory pathways of epigenetics [13]. Zhang et al. revealed that the differentially expressed lncRNAs between normal sperm and sperm from asthenozoospermic patients showed involvement of lncRNAs in different processes of spermatogenesis, as determined using gene ontology and pathway analysis [56]. The lncRNA Mrhl (Meiotic recombination hot spot1 locus) interacts with the p68 (DEAD-box helicase 5) protein and binds to the SOX8 (SRY-box 8) promoter to inhibit the Wnt signaling pathway in mouse spermatogonial cells to ensure normal sperm production [66]. The apoptosis and proliferation of male germ stem cells are regulated by lncRNA H19 through the IGF-1 signaling pathway [5]. The suppressing effect of lncRNA Gm2044 is mediated by the miR-202-Rbfox2 molecular signaling pathway in the proliferation of human testicular embryonic carcinoma cells. This provides new clues to understand male reproduction and the importance of lncRNAs in the miR-202-Rbfox2 molecular signaling pathway in the pathogenesis and treatment of male infertility [92]. Wichman et al. postulated the influence of some lncRNAs in fertility and spermatogenesis and explored the dysregulation of specific lncRNAs, which is a novel mechanism indicating fertility and low sperm quality, helping to identify new therapeutic strategies and biomarkers [54].
Conclusions and future directions
Advances in transcriptomic studies, especially in RNA sequencing, have increased the identification of lncRNAs. Recently developed computational methods for lncRNA identification and functional prediction have increased our knowledge of the various functions of lncRNAs. This has helped us to understand the complex lncRNA associated processes in various systems, lncRNAs’ molecular functions, and their mechanisms. LncRNAs have an important function in mammalian spermatogenesis; however, the specific roles of only a few lncRNAs have been determined. Therefore, it remains challenging to develop accurate and efficient methods to characterize lncRNAs and lacks detailed information about the specific roles of lncRNAs and the interaction between lncRNAs, genes, proteins, and other molecules in specific tissues and cells. Therefore, developing more powerful computational methods and comprehensive databases, and performing further experimental studies are required to determine the molecular functions, mechanisms, specific expression, and tissue specificity of lncRNAs in different cellular developmental processes. Future studies combining different approaches in lncRNA research are likely to provide a comprehensive understanding and exciting insights into this rapidly evolving field of study in complex biological processes.
The localization of lncRNAs in the vicinity of protein-coding genes and the existence of conserved lncRNAs indicate closer interactions and interrelated functions of lncRNAs in the mammalian genome. The conserved lncRNAs might also represent new prognostic biomarkers and novel diagnostic options to design therapeutic drugs. Recent advances in nucleic acid drug development have also started to provide successful solutions to male sterility problems, male contraceptives, and will allow the exploration of lncRNAs viable targets to combat male infertility, pathogenesis [92], and other pathologies.
Currently, rapid advances are being made in lncRNA pathway studies and understanding their molecular functions [5, 92, 93]. Researchers have developed alternative methods to silence the genes; however, they have failed to develop effective methods to express therapeutic genes, which remains challenging. LncRNAs could provide the best option to target a defined subset of genes found within their vicinity, in either proximal or distal positions. Although such advances in therapeutic technologies are encouraging, there are several challenges that must be addressed before their clinical applications. LncRNAs are considered to be suitable for therapeutic targets because of their presence in specifically defined developmental stages and cell-specific expressions. Therefore, further research should focus on the overall expression, off-target effects, the status of the immune response, and other issues related to lncRNAs.
Availability of data and materials
Not applicable.
Abbreviations
- AS:
-
Type A spermatogonia
- Cas9:
-
CRISPR-associated protein-9 nuclease
- cDNA:
-
Complementary DNA
- ceRNAs:
-
Competing endogenous RNAs
- CHIP-seq:
-
Chromatin immunoprecipitation sequencing
- CONC:
-
Coding, noncoding
- CRISPR:
-
Clustered regularly interspaced short palindromic
- DMRT1:
-
Doublesex and mab-3 related transcription factor 1
- DNA:
-
Deoxyribonucleic acid
- EST:
-
Expressed sequence tag
- Gas5:
-
Growth arrest-specific 5
- HOTAIR:
-
Hox transcript antisense intergenic RNA
- IGF:
-
Insulin-like growth factor
- ISH:
-
In situ hybridization
- kb:
-
Kilo base
- LncRNA:
-
Long noncoding ribonucleic acid
- LncRNA-Tcam1 :
-
LncRNA-testicular cell adhesion molecule 1
- LTR:
-
Long terminal repeat
- Malat1 :
-
Metastasis-associated lung adenocarcinoma transcript 1
- miRNA:
-
microRNA
- Mrh1 :
-
Meiotic recombination hot spot1 locus
- mRNA:
-
messenger RNA
- MSCI:
-
Meiotic sex chromosome inactivation
- Neat1 :
-
Nuclear enriched abundant transcript
- ORF:
-
Open reading frame
- RNA:
-
Ribonucleic acid
- rRNA:
-
Ribosomal RNA
- RS:
-
Round spermatids
- SAGE:
-
Serial analysis of gene expression
- SP:
-
Pachytene spermatocytes
- SSC:
-
Spermatogonial stem cells
- SVM:
-
Support vector machine
- TERRA :
-
Telomeric repeat-containing RNA
- tRNA:
-
Transfer RNA
- Tslrn1 :
-
Testis specific lncRNA1
- Tsx:
-
Testis-specific X-linked
- UTR:
-
Untranslated region
- Xist:
-
X-inactive specific transcripts
References
Mukherjee A, Koli S, Reddy KVR. Regulatory non-coding transcripts in spermatogenesis: Shedding light on “dark matter”. Andrology. 2014;2:360–9.
Bettegowda A, Wilkinson MF. Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1637–51.
Donnell LO, Robertson KM, Jones ME, Simpson ER, Henry P. Estrogen and Spermatogenesis*. Endocr Rev. 2001;22:289–318.
Agbor VA, Tao S, Lei N, Heckert LL. A Wt1-Dmrt1 transgene restores DMRT1 to Sertoli cells of Dmrt1−/− testes: a novel model of DMRT1-deficient germ cells. Biol Reprod. 2013;88:1–15.
Lei Q, Pan Q, Li N, Zhou Z, Zhang J, He X, et al. H19 regulates the proliferation of bovine male germline stem cells via IGF-1 signaling pathway. J Cell Physiol. 2018;234:915–26.
Wen K, Yang L, Xiong T, Di C, Ma D, Wu M, et al. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res. 2016;26:1233–44.
Liu K-S, Mao X-D, Pan F, Gao L-J, Ling X-F. Role of related regulatory Long noncoding RNAs on mammalian spermatogenesis. Reprod Dev Med. 2017;1:18–22.
Lü M, Tian H, Cao Y, He X, Chen L, Song X, et al. Downregulation of miR-320a/383-sponge-like long non- coding RNA NLC1-C (narcolepsy candidate-region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death Dis. 2015;6:e1960.
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61 Elsevier Ltd.
Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:1–16.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;457:223–7 Macmillan Publishers Limited.
Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58 Elsevier Ltd.
Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7.
Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. 2011;43:904–14 Elsevier Inc.
Okamura K. Diversity of animal small RNA pathways and their biological. Wiley Interdiscip Rev RNA. 2012;3:351–68.
Liu K, Mao X, Chen Y, Li T, Ton H. Regulatory role of long non-coding RNAs during reproductive disease. Am J Transl Res. 2018;10:1–12.
Li L, Wang M, Wu X, Geng L, Xue Y, Wei X, et al. A long non-coding RNA interacts with Gfra1 and maintains survival of mouse spermatogonial stem cells. Cell Death Dis. 2016;7:e2140.
Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–21.
Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–23.
Zhang XJ, Liu ML, Jia MC. Regulation of gene expression during spermatogenesis at transcriptional level. Hered. 2011;33:1300–7.
Sun J, Lin Y, Wu J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS One. 2013;8:e75750.
Di C, Yuan J, Wu Y, Li J, Lin H, Hu L, et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014;80:848–61.
Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–5 Macmillan Publishers Limited.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41 Elsevier Inc.
Derrien T, Johnson R, Bussotti G, Frazer KA. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89.
Nam JW, Bartel DP. Long non-coding RNAs in C. elegans. Genome Res. 2012;22:2529–40.
Tsai M, Spitale RC, Chang HY. Long Intergenic noncoding RNAs: new links in Cancer progression Long Intergenic noncoding RNAs: new links in Cancer. Am Assoc Cancer Res. 2011;71:3–7.
Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS. Long non-coding RNA: its evolutionary relics and biological implications in mammals: a review. J Anim Sci Technol. 2018;60:1–10.
Khachane AN, Harrison PM. Mining mammalian transcript data for functional Long. PLoS One. 2010;5:e10316.
Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–43.
De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi K, et al. A large fraction of Extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384.
Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at Most promoters in human cells. Cell. 2007;130:77–88.
Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:1–10.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:1–16.
Hu K, Zhang J, Liang M. LncRNA AK015322 promotes proliferation of spermatogonial stem cell C18-4 by acting as a decoy for microRNA-19b-3p. Vitro Cell Dev Biol. 2016;1:277–84.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23.
Iwakiri J, Hamada M, Asai K. Bioinformatics tools for lncRNA research. Biochim Biophys Acta. Biochim Biophys Acta. 2016;1859:23–30 Elsevier B.V.
Zhou B, Zhao H, Yu J, Guo C, Dou X, Song F, et al. EVLncRNAs : a manually curated database for long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2018;46:100–5 Oxford University Press.
Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, et al. Lncipedia 5: towards a reference set of human long non-coding rnas. Nucleic Acids Res. 2019;47:D135–9.
Bu D, Yu K, Sun S, Xie C, Skogerbø G, Miao R, et al. Noncode v3. 0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:210–5.
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47:D1034–7.
Chakraborty S, Deb A, Maji RK, Saha S, Ghosh Z. LncRBase: an enriched resource for lncRNA information. PLoS One. 2014;9:e108010.
Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. LncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:146–51.
Pian C, Zhang G, Tu T, Ma X, Li F. LncCeRBase: a database of experimentally validated human competing endogenous long non-coding RNAs. Database. 2018;2018:1–4.
Petrov AI, Kay SJE, Kalvari I, Howe KL, Gray KA, Bruford EA, et al. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017;45:D128–34.
Bhartiya D, Pal K, Ghosh S, Kapoor S, Jalali S, Panwar B, et al. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database. 2013;2013:1–7.
Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:122–6.
Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell. 2013;152:1298–307 Elsevier Inc.
Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7.
Flynn RA, Chang HY. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Stem Cell. 2014;14:752–61 Elsevier Inc.
Bao J, Wu J, Schuster AS, Hennig GW, Yan W. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male Germline1. Biol Reprod. 2013;89:1–12.
Song X, Kyi-Tha-Thu C, Takizawa T, Naing BT, Takizawa T. 1700108J01Rik and 1700101O22Rik are mouse testis-specific long non-coding RNAs. Histochem Cell Biol. 2018;149:517–27.
Wichman L, Somasundaram S, Breindel C, Valerio DM, Mccarrey JR, Hodges CA, et al. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility †. Biol Reprod. 2017;97:313–23.
Dai YB, Lin Y, Song N, Sun F. LncRNA4667 is dispensable for spermatogenesis and fertility in mice. Reprod Dev Med. 2019;3:18–23.
Zhang X, Zhang P, Song D, Xiong S, Zhang H, Fu J, et al. Expression profiles and characteristics of human lncRNA in normal and asthenozoospermia sperm†. Biol Reprod. 2018;100:982–93.
Zhang L, Liu Z, Li X, Zhang P, Wang J, Zhu D, et al. Low long non-coding RNA HOTAIR expression is associated with down-regulation of Nrf2 in the spermatozoa of patients with asthenozoospermia or oligoasthenozoospermia. Int J Clin Exp Pathol. 2015;8:14198–205.
Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2015;44:D196–202.
Liang M, Li W, Tian H, Hu T, Wang L, Lin Y, et al. Sequential expression of long noncoding RNA as mRNA gene expression in specific stages of mouse spermatogenesis. Sci Rep. 2014;4:1–6.
Lee T, Xiao A, Rennert OM. Identifi cation of Novel Long Noncoding RNA Transcripts in Male Germ Cells. Methods Mol Biol. 2012;825:105–14.
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208.
Yamada A, Yu P, Lin W, Okugawa Y, Boland CR, Goel A. A RNA-sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci Rep. 2018;8:2–11 Springer US.
Clark MB, Mercer TR, Bussotti G, Leonardi T, Haynes KR, Crawford J, et al. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods. 2015;12:339–42.
Rathke C, Barckmann B, Burkhard S, Jayaramaiah-raja S, Roote J, Renkawitz-pohl R. Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur J Cell Biol Elsevier. 2010;89:326–38.
Zhang L, Lu H, Xin D, Cheng H, Zhou R. A novel ncRNA gene from mouse chromosome 5 trans-splices with Dmrt1 on chromosome 19. Biochem Biophys Res Commun. 2010;400:696–700 Elsevier Inc.
Arun G, Akhade VS, Donakonda S, Rao MRS. Mrhl RNA, a Long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse Spermatogonial cells. Mol Cell Biol. 2012;32:3140–52.
Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116:459–66.
Gibb EA, Vucic EA, Enfield KSS, Stewart GL, Lonergan KM, Jennifer Y, et al. Human Cancer Long non-coding RNA Transcriptomes. PLoS One. 2011;6:1–10.
Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, et al. Clusters of internally primed transcripts reveal novel Long noncoding RNAs. PLoS Genet. 2006;2:e37.
Huang W, Long N, Khatib H. Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim Genet. 2012;43:674–82.
Pauli A, Valen E, Lin MF, Pauli A, Valen E, Lin MF, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–91.
Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322:750–6.
Okazaki Y, Furuno M, Kasukawa T, Schonbach C, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Hasegawa Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–73.
Gruber AR, Bernhart SH, Hofacker IL, Washietl S. Strategies for measuring evolutionary conservation of RNA secondary structures. BMC Bioinformatics. 2008;9:122.
Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:275–82.
Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:251–62.
Liu J, Gough J, Rost B. Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet. 2006;2:e29.
Hu K, Li L, Liao Y, Liang M. LncRNA Gm2044 highly expresses in spermatocyte and inhibits Utf1 translation by interacting with Utf1 mRNA. Genes Genomics. 2018;40:781–7.
Akhade VS, Arun G, Donakonda S, Rao MRS. Genome wide chromatin occupancy of mrhl RNA and its role in gene regulation in mouse spermatogonial cells. RNA Biol. 2014;11:1262–79.
Liang M, Li W, Tian H, Hu T, Wang L, Lin Y, et al. RNA as mRNA gene expression in. Sci Rep. 2014;4:1–6.
Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7:1–14.
Satoh Y, Takei N, Kawamura S, Takahashi N, Kotani T, Atsushi P. A novel testis-specific long noncoding RNA , Tesra , activates the Prss42 / Tessp-2 gene during mouse spermatogenesis †. Biol Reprod. 2019;100:833–48.
Gascoigne DK, Cheetham SW, Cattenoz PB, Clark MB, Amaral PP, Taft RJ, et al. Sequence analysis pinstripe : a suite of programs for integrating transcriptomic and proteomic datasets identifies novel proteins and improves differentiation of protein-coding and non-coding genes. Bioinformatics. 2012;28:3042–50.
Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–40.
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Gerd A, et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. 2014;494:497–501.
Melo CA, Drost J, Wijchers PJ, Van De Werken H, De Wit E, Melo A, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:1–12.
Ørom UA, Shiekhattar R. Long Noncoding RNAs Usher in a New Era in the Biology of Enhancers. Cell. 2013;154:1190–3 Elsevier Inc.
Ni MJ, Hu ZH, Liu Q, Liu MF, Lu MH, et al. Identification and Characterization of a Novel Non- Coding RNA Involved in Sperm Maturation. PLoS One. 2011;6:e26053.
Akhade VS, Dighe SN, Kataruka S, Rao MRS. Mechanism of Wnt signaling induced down regulation of mrhl long non-coding RNA in mouse spermatogonial cells. Nucleic Acids Res. 2016;44:387–401.
West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, et al. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol Cell. 2014;55:791–802 Elsevier Inc.
Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403 Elsevier Inc.
Liang M, Hu K, He C, Zhou J, Liao Y. Upregulated lncRNA Gm2044 inhibits male germ cell development by acting as miR-202 host gene. Animal Cells Syst. 2019;23:128–34.
Lavorgna G, Vago R, Sarmini M, Montorsi F, Salonia A, Bellone M. Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res. 2016;110:131–8 Elsevier Ltd.
Acknowledgments
The authors are grateful to the native English-speaking scientists of “Elixigen Company (Huntington Beach, California)” for proofreading the article.
Funding
This work was supported by the National Natural Science Foundation of China (3187131067), the Agricultural Science and Technology Innovation Program (ASTIP-2016-IAS-06 and CAAS-XTCX2016010), and the China Agriculture Research System (CARS-36).
Author information
Authors and Affiliations
Contributions
BW, SJ, and HB conceived the idea and outlined the sketch. BW, and SJ performed the literature search, wrote the initial manuscript of each section, and constructed the figures and tables. JM, and HB conceptualized the review, literature search, and the writing and editing of the manuscript and figure legends. XG, SU and HY modified the manuscript. BW and SJ contributed equally to this work, and all authors read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sahlu, B.W., Zhao, S., Wang, X. et al. Long noncoding RNAs: new insights in modulating mammalian spermatogenesis. J Animal Sci Biotechnol 11, 16 (2020). https://doi.org/10.1186/s40104-019-0424-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-019-0424-8