Abstract
Background
Clostridium perfringens is the main etiological agent of necrotic enteritis. Lactobacilli show beneficial effects on intestinal health in infectious disease, but the protective functions of lactobacilli in C. perfringens-infected chickens are scarcely described. This study examined the effects of Lactobacillus acidophilus (L. acidophilus) on the growth performance and intestinal health of broiler chickens challenged with Clostridium perfringens (C. perfringens) over a 28-day period. Using a 2 × 2 factorial arrangement of treatments, a total of 308 1-day-old male Arbor Acres broiler chicks were included to investigate the effects of Lactobacillus acidophilus (L. acidophilus) on the growth performance and intestinal health of broiler chickens challenged with Clostridium perfringens (C. perfringens) during a 28-day trial.
Results
During infection (d 14–21), C. perfringens challenge decreased the average daily gain (P < 0.05), and increased feed conversion ratio and the mortality rate (P < 0.05). However, dietary supplementation with L. acidophilus increased the body weight of C. perfringens-infected broilers on d 21 (P < 0.05), and tended to decrease the mortality (P = 0.061). C. perfringens challenge decreased the villus height (P < 0.05), the ratio of villus height to crypt depth (P < 0.05) and OCLN (occludin) mRNA expression (P < 0.05), and increased the pro-inflammatory cytokine expression in the spleen and jejunum, the intestinal populations of C. perfringens and Escherichia (P < 0.05), and the serum content of endotoxin (P < 0.05), regardless of L. acidophilus supplementation. In contrast, dietary L. acidophilus reducedthe intestinal lesion score of challenged broilers (P < 0.05), the mRNA expression of pro-inflammatory cytokines, ileal populations of Escherichia and serum endotoxin content (P < 0.05), but increased the intestinal Lactobacillus populations (P < 0.05), irrespective of C. perfringens challenge.
Conclusion
Dietary addition of L. acidophilus could improve the intestinal health and reduce the mortality of broilers suffering from necrotic enteritis.
Similar content being viewed by others
Background
Necrotic enteritis (NE) is a wide-spread poultry disease costing the global poultry industry approximately 2 billion U.S. dollars each year [1]. Clostridium perfringens (C. perfringens) type A is the main etiological agent of NE, and causes two forms of disease: a clinical form, with a sudden increase in flock mortality, often without premonitory signs, and a subclinical form, causing sub-optimal growth performance [2]. In-feed antibiotic growth promoters have been used to control NE; however, many countries forbid the dietary use of antimicrobials because of public concern over the emergence of antibiotic-resistant bacteria [3]. Outbreaks of NE have been increasing due to the withdrawal of in-feed antibiotics [4], indicating a threat to human food security. In recent years, probiotics have been used as an alternative to antibiotics. As probiotics, Lactobicillus species have been used to improve the intestinal health and growth performance of poultry. Lactobacilli produce bacteriostatic bacteriocin-like compounds [5] as well as acids, such as lactic acid, which decreases the pH of the gut. Competitive exclusion and antagonism have also been proposed as mechanisms by which lactobacilli species prevent the proliferation of pathogenic bacteria and regulate the intestinal flora [6,7,8]. Previous studies have shown that Lactobacillus acidophilus (L. acidophilus) can inhibit the pathogens [9, 10] and modulate the immunity [11]. However, it is unclear whether dietary L. acidophilus supplementation could improve the intestinal health and growth performance of broilers in a NE model. Therefore, we conducted a NE model-based feeding trial to investigate the effects of L. acidophilus on the intestinal permeability andhistomorphology, cytokine mRNA expression, and microbiota of broiler chickens challenged with C. perfringens.
Methods
Animals and experimental design
A total of 308 1-day-old male Arbor Acres broilers were used to study the effects of additives (without/with L. acidophilus), pathogen challenge (without/with C. perfringens challenge), and their interactive effects. A 2 × 2 factorial arrangement was used, with a completely randomized experimental design. All newly hatched healthy birds were weighed and randomly assigned into one of four treatment groups, with seven replicates in each group and 11 birds per replicate. The four groups consisted ofan untreated control (CTL), a L. acidophilus only supplementation group (LA), a C. perfringens only challenge group (CLG), and a C. perfringens challenge group supplemented with L. acidophilus (CLG + LA). All birds were housed in the cages (100 cm × 100 cm), and were offered free access to the feed and water throughout the 28-day trial.
Diets and L. acidophilus supplementation
A corn-soybean-meal basal diet in mash form was formulated to meet the nutrient requirements of chickens, as per the recommended feeding standards for broilers in China (NY/T 2004) (Table 1). The ingredients of basal diet were mixed in the same batch to ensure that the experimental diets were identical in composition. A probiotic formulation of L. acidophilus LAP5 (Synlac Material Technology Co., Nanjing, China) was added to the basal diet at 40 mg/kg, providing 4.0 × 106 cfu/kg of diet. Firstly, we calculated the exact amount of bacteria and basal diet that were needed to produce the experimental diets; secondly, the bacteria were diluted with a small amount of basal diet; thirdly, it would be mixed with the remaining basal diet using the shovel. The experimental diets were produced manually every 4 d.
C. perfringens challenge
The C. perfringens challenge method used in this study was developed by Dahiya et al. [12], and modified by Liu et al. [13]. Briefly, a chicken C. perfringens type A field strain (CVCC 2030), originally isolated from a clinical case of NE, was obtained from the China Veterinary Culture Collection Center (Beijing, China). C. perfringens was cultured anaerobically on tryptose-sulfite-cycloserine agar for 18 h at 37 °C, and then aseptically inoculated into cooked meat medium and incubated anaerobically for 8 h at 37 °C. Birds in the challenge groups were orally gavaged once daily with actively growing C. perfringens (2.0 × 108 cfu/mL, 1.0 mL per bird) during d 14–20, while the non-challenged birds were gavaged with the same volume of sterilized cooked meat medium.
Sample collection
On d 21, one bird in each replicate was randomly selected and humanly euthanized to collect the blood, fresh digesta in the ileum (defined as the region between Meckel’s diverticulum and 2 cm cranial to the ileo-caecal junction) and cecum, spleen, and segments in the middle of jejunum (defined as the region from the end of duodenum to the Meckel’s diverticulum). The blood was collected aseptically from the wing vein into normal vacutainers. Serum was then obtained from the blood samples and stored at − 20 °C. Spleen and intestinal segments for mRNA isolation were frozen in liquid nitrogen. Fresh ileal digesta (collected from the segments between ileum midpoint and 2 cm proximal to the ileocecal junction) and cecal digesta were collected aseptically and stored at − 70 °C to determine the quantitation of the C. perfringens, Escherichia and Lactobacillus species populations.
Growth performance
Body weight (BW) and feed intake (FI) for each replicate were measured on d 14, 21 and 28. Average daily gain (ADG), average daily feed intake (ADFI), and the feed conversion ratio (FCR) were calculated during the different periods (d 1–14, d 14–21 and d 21–28).
Intestinal lesion score
The small intestine of each bird was excised and subjected to scoring for NE lesions on a scale from 0 to 4, as described by Liu et al. [13]. Briefly, 0 = normal intestinal appearance; 0.5 = severely congested serosa and mesentery engorged with blood; 1 = thin-walled and friable intestines with small red petechiae; 2 = focal necrosis, grey appearance and small amounts of gas production; 3 = sizable patches of necrosis, gas-filled intestine, and small flecks of blood; and 4 = severe extensive necrosis, marked hemorrhage, and large amounts of gas in the intestine.
Intestinal histomorphology
A segment of ileum from each bird was fixed in 4% paraformaldehyde immediately after sacrifice and then embedded in paraffin. Transverse 5-μm sections were stained with hematoxylin and eosin, and then histomorphometrically examined using an Olympus optical microscope and ProgRes CapturePro software (version 2.7; Jenoptik, Jena, Germany). Ten villi were measured in each section and only complete and vertically oriented villi were measured. Villus height was measured from the tip of the villus to the crypt opening and the associate crypt depth was measured from the base of the crypt to the level of the crypt opening. Then the average values of villus height and crypt depth in each section were calculated, respectively. The ratio of villus height to relative crypt depth was calculated from these measurements.
Serum endotoxin content
The barrier integrity and function were evaluated using an indirect method by measuring the serum endotoxin levels. A Quantitative Chromogenic End-point Tachypleus Amebocyte Lysate Endotoxin Detection Kit was used, following the manufacturer’s instructions (Xiamen TAL Experimental Plant Co., Fujian, China).
Gene expression analysis
Total RNA isolation was carried out by using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. The concentration and purity of total RNA was monitored using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). One microgram of total RNA was reverse transcribed by a reverse transcription kit (Takara Bio Inc.) according to the manufacturer’s instructions. Reverse transcription was performed at 42 °C for 2 min, 37 °C for 15 min, followed by heat inactivation for 5 s at 85 °C. All of the cDNA preparations were stored frozen at 20 °C until further use. On d 21, the mRNA expression of IL-1β, IL-8, interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and IL-10 in the spleen and jejunum and mucin 2 (MUC2), claudin 1 (CLDN1), occludin (OCLN) and zonula occludens 1 (ZO-1) in the jejunum were measured by quantitative real-time PCR (qRT-PCR) analysis. Gene specific primer sequences are shown in Table 2. The qRT-PCR assay was performed using a 7500-fluorescence detection system (Applied Biosystems, Foster City, California) and a commercial SYBR-Green PCR kit (Takara Bio Inc., Ostu, Japan). The following thermal cycler conditions were used: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 34 s. At the end of each run, melting curve analysis and subsequent agarose gel electrophoresis of the PCR products were carried out to confirm the amplification specificity. The β-actin was used as the housekeeping gene, and the data of relative gene expression were analyzed using the 2−ΔΔCt method as previously described (Livak and Schmittgen, [14]). The amplifying efficiency of the qRT-PCR primers for each target gene is between 90% and 110%.
Ileal and cecal microbiota enumeration
The populations of C. perfringens, Escherichia subgroup, and Lactobacillus subgroup species in the digesta were detected by absolute qRT-PCR, as described previously [15, 16], with some modification. Briefly, genomic DNA was isolated from about 200 mg of digesta from the ileum and caecum using a QIAamp DNA Stool Mini Kit (Qiagen Inc., Valencia, CA). Extracted DNA was stored at − 70 °C until analysis. Standard curves for qRT-PCR were prepared by normal PCR amplification using DNA extracted from pure bacterial cultures to produce a high concentration of the target DNA. Competent Escherichia coli DH5α (Takara Bio Inc., Japan) were used to generate plasmid standards. The PCR products were purified using a PCR purification kit (Biomed Gene Technologies, Beijing, China), and then cloned into pCR2.1 using a TA cloning kit (Invitrogen Corporation, Carlsbad, CA), as per the manufacturer’s protocol. Purified insert-containing plasmids were quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA), and the number of target gene copies was calculated using the following formula, as described previously [17]:
Standard curve was constructed by the ten-fold serial dilutions of plasmid DNA. Genomic DNA from ileal and cecal samples was used as a template for absolute qRT-PCR using a 7500 fluorescence detection system (Applied Biosystems, Foster City, CA) according to optimized PCR protocols (SYBR-Premix Ex Taq, Takara Bio Inc., Japan). The qRT-PCR primers were the same as used for normal PCR amplification (Table 3). The data were presented as log10 gene copies/g of intestinal digesta.
Statistical analysis
All data were analyzed with SPSS version 20.0 (SPSS Inc., Chicago, IL). A significance level of 0.05 was used. The data of mortality and intestinal lesion score were analyzed by one-way ANOVA, and were subjected to grouped table of the GraphPad Prism 5 (GraphPad Software, Inc., CA, USA). Results in figures were represented as mean ± SE of 7 replicates, with 11 birds per replicate. Other data were analyzed by two-factorial analysis of variance to examine the main effects of probiotic and challenge, and their interaction using general linear model procedure SPSS 20.0. When a significant interaction between the main effects was observed, Duncan’s multiple comparison was used to compare the differences among the four groups. Results in the tables were given as the mean and pooled SEM.
Results
Growth performance and mortality
The growth performance of broiler chickens was not significantly affected by dietary L. acidophilus addition during the d 0–14 (P > 0.05) (data not shown). However, as shown in Table 4, C. perfringens challenge decreased the ADG (P < 0.05) and increased the FCR (P < 0.05) of broilers during d 14–21. An interaction between the effects C. perfringens and L. acidophilus treatment on BW of broilers was observed on d 21 (P < 0.05), and L. acidophilus treatment significantly increased the BW of broilers when challenged (P < 0.05). No deaths were observed in non-challenged broilers (CTL and LA groups) during the experimental period, while C. perfringens challenge significantly increased the mortality rate (P < 0.05) of broilers from d 14 to 21, and L. acidophilus addition tended to decrease the mortality rate (P = 0.061) (Fig. 1). The growth performance of broilers during d 21–28 was not significantly affected by C. perfringens challenge or L. acidophilus supplementation (P > 0.05, data not shown).
Intestinal lesion score
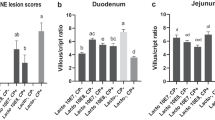
In this study, no intestinal lesions were observed in the non-challenged broiler chickens. Most C. perfringens-challenged birds exhibited congested mucosa and focal haemorrhagic lesions in the jejunum and ballooning in the small intestine was observed in some challenged birds. However, L. acidophilus supplementation in the diet significantly decreased the intestinal lesion score (P < 0.05) of challenged birds (Fig. 2).
Intestinal histomorphology
Clostridium perfringens challenge significantly decreased the villus height (P < 0.05) and the ratio of villus height to crypt depth ratio (P < 0.05), and tended to increase the crypt depth (P = 0.059, Table 5). L. acidophilus treatment tended to increase the villus height (P = 0.057) and significantly increased crypt depth (P < 0.05) in the both non-challenged and challenged birds.
Serum endotoxin content
The serum endotoxin was measured to estimate intestinal barrier integrity and function (Table 6). Clostridium perfringens challenge and L. acidophilus treatment had interactive effects on the levels of serum endotoxin (P < 0.05). The serum endotoxin content was the highest in the birds of CLG group, and was the lowest in birds of LA group. L. acidophilus supplementation significantly decreased the serum endotoxin content (P < 0.05) indespite of C. perfringens challenge.
MUC2, CLDN1, OCLN and ZO-1 mRNA expression
As shown in Table 7, C. perfringens infection significantly decreased the OCLN mRNA expression (P < 0.05), while L. acidophilus treatment tended to decrease MUC2 mRNA expression in the jejunum (P = 0.082). The relative mRNA levels of CLDN1 and ZO-1 expression in the jejunum were not significantly affected by C. perfringens infection or L. acidophilus treatment (P > 0.05).
Cytokine mRNA expression
The expression of IL-1 β, IFN-γ, and TNF-α in the spleen was significantly (P < 0.05) up-regulated by C. perfringens challenge (Table 8), while L. acidophilus treatment alone significantly (P < 0.05) down-regulated the expression of IL-1 β and TNF-α. Interestingly, interactive effects on the IL-8 mRNA expression (P < 0.05) in the spleen were observed between C. perfringens challenge and L. acidophilus treatment. IL-8 mRNA expression in the spleen of broilers in CLG treatment was significantly higher than that in non-challenged birds (P < 0.05). However, the IL-8 mRNA expression in the spleen was significantly (P < 0.05) decreased in the challenged broilers fed L. acidophilus-supplemented diet.
As shown in Table 9, C. perfringens challenge increased the mRNA expression of IL-1 β (P < 0.05) in the jejunum. Conversely, L. acidophilus treatment significantly decreased the expression of IL-1 β (P < 0.05), and tended to decrease TNF-α expression in the jejunum of broilers at d 21 (P = 0.074). Interactive effects on the expression of IL-8 (P < 0.05), IFN-γ (P < 0.05), and IL-10 (P < 0.05) in the jejunum were observed between C. perfringens challenge and L. acidophilus treatment. Furthermore, L. acidophilus treatment decreased the expression of IL-8 (P < 0.05), IFN-γ (P < 0.05), and IL-10 (P < 0.05) in the jejunum of challenged broilers.
Enumerations of ileal and cecal microbiota
The populations of C. perfringens (P < 0.05), Lactobacillus (P < 0.05), and Escherichia (P < 0.05) in the ileum and populations of C. perfringens (P < 0.05) and Escherichia (P < 0.05) in the cecum were significantly increased by C. perfringens challenge (Table 10), and Lactobacillus number in the cecum of broilers at d 21 was tended to be increased by challenge (P = 0.098). L. acidophilus supplementation into the diet significantly (P < 0.05) increased the Lactobacillus population in the both ileum and cecum, and significantly (P < 0.05) decreased the Escherichia population in the ileum of broilers at d 21.
Discussion
The current study highlighted the significance of C. perfringens infection in thepoultry production. The mortality rate of C. perfringens-challenged birds was the highest out of all groups examined in this study, but was lower than the 36% mortality rate reported in a previous clinical NE study [18], which might indicate the strain specificity of C. perfringens. Pathogen challenge also increased the intestinal lesion score and decreased the intestinal villus height and the ration of villus height to crypt depth ratio in the present study. Intestinal NE lesions and mucosal atrophy greatly compromises epithelial permeability and mucosal barrier function [19], resulting in bacteria translocation to the liver, spleen, and blood [13]. This is reflected in increased levels of blood endotoxin produced by Gram-negative bacteria. Accordingly, we observed that the serum endotoxin content of broilers in the infected groups was higher than that in the uninfected groups.
Lactobacilli can inhibit pathogen growth and enhance poultry productivity. However, there are few reports on the effects of lactobacilli on NE-infected broilers. Geier et al. [18] reported that L. johnsonii appeared to reduce intestinal lesion score and the ratio of feed to weightgain, but did not decrease the mortality of NE-infected broilers. L. fermentum 1.2029 reduced the NE lesion severity and attenuated intestinal damage caused by C. perfringens challenge [20]. In the current study, L. acidophilus supplementation in the diet improved growth performance, reduced the mortality rates, and attenuated cellular damage of challenged broilers. Italso increased the jejunal crypt depth, which might aid the intestinal renewal and the recovery of NE-infected broiler chickens.
Tight junctions are the most important components of the intestinal epithelial cell barrier, which protects the host from intestinal pathogens and prevents macromolecular transmission [21]. Tight junctions are formed by several types of proteins, including OCLN, claudins, junctional adhension molecules, and zonula occludens (ZO) proteins [22]. To date, only claudin-1, claudin-2, claudin-3, claudin-5, claudin-16, ZO-1, ZO-2, and OCLN have been reported in poultry [23,24,25,26]. In the present study, C. perfringens challenge decreased the OCLN mRNA expression, but did not significantly affect the expression of CLDN1 or ZO-1 in the jejunum. This finding was consistent with a previous report [13], which demonstrated that OCLN mRNA expression in the jejunum and ileum was down-regulated by C. perfringens infection. Du et al. [27] also reported that the CLDN1 expression in the ileum was decreased by C. perfringens challenge. However, in the current study, L. acidophilus treatment did not show significant effect on the expression of CLDN1, OCLN or ZO-1, but did decrease MUC2 mRNA expression. The specific strain of L. acidophilus used in this study might not benefit the host by strengthening the tight junctions.
Many types of bacteria have been shown to co-exist in case of NE, and one of the largest populations is E. coli [28], although Lactobacillus species arepresent as well [13, 29]. In the current study, C. perfringens challenge increased the populations of C. perfringens and Escherichia in the ileum and cecum, and also increased ileal Lactobacillus counts, which is consistent with previous reports [13, 16]. Lactobacilli can prevent the growth of C. perfringens in vitro [30]. It was also demonstrated that some Lactobacillus strains have demonstrated the capacity to prevent C. perfringens colonization and NE outbreak in vivo. Addition of Lactobacillus sp. NO. I-2673 to drinking water decreased the cecal C. perfringens populations of healthy chickens [31], while La Ragione et al. [32] reported that L. johnsonii FI9785 prevented C. perfringens colonization in pathogen-free broilers. However, Geier et al. [18] observed that L. johnsonii did not exhibit any an-ticlostridial properties in vivo. The differences among these reports may indicate strain specificity of L. johnsonii fighting against C. perfringens. In this study, supplementation of L. acidophilus in the diet increased Lactobacillus numbers in the ileum and cecum, and decreased the ileal Escherichia counts, which might help counteract the increased serum endotoxin content induced by C. perfringens challenge. However, L. acidophilus treatment did not significantly affect the intestinal C. perfringens counts in the current study. In contrast, Fukata et al. [33] reported that L. acidophilus could inhibit the toxins production of C. perfringens, which might help explain the beneficial effect after probiotic addition.
The host immune responses are triggered after birds are infected by a pathogen. If the pathogen-induced inflammatory responses get out of control, tissue damage occurs, accompanied by suboptimal growth performance, which was also observed in the current study. Previous studies have demonstrated that theexpression of IFN-γ, IL-1β, IL-4, and IL-10 was increased in the intestines of NE-infected broilers compared with uninfected controls [34, 35]. However, the immune responses to C. perfringens infection may differ due to the varieties of broiersand vary in different immune organs. Hong et al. [36] observed that NE-infected broilers showed up-regulated transcription of the pro-inflammatory cytokine-encoding genes IL-1β, IL-6, IL-17F, and TNFSF15 in the spleen and TNFSF15 in the intestine, but the expression of IL-17F was only increased in the intestines of Ross chickens, and was not significantly affected in the intestines of Cobb chickens. In addition, Lee et al. [37] reported that IL-8, LITAF, TNFSF15, IL-17A and IL-17F transcript levels in the jejunum of broilers were up-regulated by Eimeria maxima/C. perfringens co-infection, while only IL-17A transcripts were increased in the spleen. In this study, the treatments affected the IL-10 mRNA expression level in the jejunum, but not in the spleen. What’s more, the inflammatory responses seemed to be more intense in the jejunum than in the spleen, which might lead to gut damage, higher energy consumption and sub-optimal growth performance. The inconsistency of previous findings and present study might be due to differences of the NE infection models and broiler varieties.
Lactobacilli has been shown to regulate the transcript levels of cytokines. In the absence of pathogen challenge, L. acidophilus was effective in inducing T-helper-1 cytokines, while L. salivarius induced an even greater anti-inflammatory response in the chicken spleen and cecal tonsil cells [38]. Wang et al. [39] reported that novel L. plantarum strain P-8 increased the jejunal transcription of IFN-γ, IL-12, and IL-4 at d 14 post treatment. Those studies demonstrated that lactobacilli could increase host immunity in the absence of pathogens. However, little was known aboutthe effects of Lactobacilli on the cytokine expression in a NE model. In the present study, L. acidophilus decreased the transcription of IL-1β and TNF-α in the spleen and jejunum of broilers, irrespective of C. perfringens challenge. In addition, the decreases of IL-8 expression were observed in the spleen, while the expression levels of IL-8, IFN-γ, and IL-10 were decreased in the jejunum of NE-infected broilers fed L. acidophilus supplemented diet compared with those infected broilers fed control diet. Overall, lactobacilli may exhibit different regulatory functions when hosts were under different conditions. Thus, the decreased cytokine levels may be associated with the protective effect of L. acidophilus treatment onthe gut health of NE-infected broilers. L. acidophilus treatment ameliorated the inflammation, and therefore reduced the consumption of energy and improved gut health, as evidenced by the decreased lesion scores and increased villus height.
Conclusions
Dietary supplementation with L. acidophilus could alleviate the inflammation and intestinal impairment, improve intestinal morphology and barrier integrity, and modulate the intestinal microflora. Consequently, L. acidophilus addition benefited the intestinal health, and decreased the mortality of broilers challenged with C. perfringens.
Abbreviations
- ADFI:
-
Average daily feed intake
- ADG:
-
Average daily gain
- BW:
-
Body weight
- C. perfringens :
-
Clostridium perfringens
- CLDN1:
-
Claudin 1
- FCR:
-
Feed conversation ratio
- FI:
-
Feed intake
- IFN-γ:
-
Interferon gamma
- L. acidophilus :
-
Lactobacillus acidophilus
- MUC2:
-
Mucin 2
- NE:
-
Necrotic enteritis
- OCLN:
-
Occludin
- TNF-α:
-
Tumor necrosis factor alpha
- ZO-1:
-
Zonula occludens 1
References
Wvd S, van der Sluis W. Clostridial enteritis is an often underestimated problem. World Poultry. 2000;16:42–3.
Van Immerseel F, Rood JI, Moore RJ, Titball RW. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 2009;17:32–6.
M'Sadeq SA, Wu S-B, Choct M, Swick RA. Heat production and energy efficiency of broilers infected with necrotic enteritis. Avian Dis. 2016;60:50–5.
Hofacre CL, Beacorn T, Collett S, Mathis G. Using competitive exclusion, mannan-oligosaccharide and other intestinal products to control necrotic enteritis. J Appl Poult Res. 2003;12:60–4.
Chateau N, Castellanos I, Deschamps AM. Distribution of pathogen inhibition in the Lactobacillus isolates of a commercial probiotic consortium. J Appl Bacteriol. 1993;74:36–40.
Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–78.
Frece J, Kos B, Svetec IK, Zgaga Z, Beganovic J, Lebos A, et al. Synbiotic effect of Lactobacillus helveticus M92 and prebiotics on the intestinal microflora and immune system of mice. J Dairy Res. 2009;76:98–104.
Liu H, Zhang J, Zhang S, Yang F, Thacker PA, Zhang G, et al. Oral Administration of Lactobacillus fermentum I5007 favors intestinal development and alters the intestinal microbiota in formula-fed piglets. J Agric Food Chem. 2014;62:860–6.
Lin C-K, Tsai H-C, Lin P-P, Tsen H-Y, Tsai C-C. Lactobacillus acidophilus LAP5 able to inhibit the Salmonella choleraesuis invasion to the human Caco-2 epithelial cell. Anaerobe. 2008;14:251–5.
Tsai CC, Hsih HY, Chiu HH, Lai YY, Liu JH, Yu B, et al. Antagonistic activity against salmonella infection in vitro and in vivo for two lactobacillus strains from swine and poultry. Int J of Food Microbiol. 2005;102:185–94.
Lin WH, Yu B, Lin CK, Hwang WZ, Tsen HY. Immune effect of heat-killed multistrain of Lactobacillus acidophilus against salmonella typhimurium invasion to mice. J Appl Microbiol. 2007;102:22–31.
Dahiya JP, Hoehler D, Wilkie DC, Van Kessel AG, Drew MD. Dietary glycine concentration affects intestinal Clostridium perfringens and lactobacilli populations in broiler chickens. Poult Sci. 2005;84:1875–85.
Liu D, Guo Y, Wang Z, Yuan J. Exogenous lysozyme influences Clostridium perfringens colonization and intestinal barrier function in broiler chickens. Avian Pathol. 2010;39:17–24.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–8.
Wise MG, Siragusa GR. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J Appl Microbiol. 2007;102:1138–49.
Du E, Gan L, Li Z, Wang W, Liu D, Guo Y. In vitro antibacterial activity of thymol and carvacrol and their effects on broiler chickens challenged with Clostridium perfringens. J Anim Sci Biotechnol. 2015;6:58.
Lee C, Kim J, Shin SG, Hwang S. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol. 2006;123:273–80.
Geier MS, Mikkelsen LL, Torok VA, Allison GE, Olnood CG, Boulianne M, et al. Comparison of alternatives to in-feed antimicrobials for the prevention of clinical necrotic enteritis. J Appl Microbiol. 2010;109:1329–38.
Sun XY, Spencer AU, Yang H, Haxhija EQ, Teitelbaum DH. Impact of caloric intake on parenteral nutrition-associated intestinal morphology and mucosal barrier function. J Parenter Enter Nutr. 2006;30:474–9.
Cao L, Yang XJ, Li ZJ, Sun FF, Wu XH, Yao JH. Reduced lesions in chickens with Clostridium perfringens-induced necrotic enteritis by Lactobacillus fermentum 1.2029. Poult Sci. 2012;91:3065–71.
Ballard ST, Hunter JH, Taylor AE. Regulation of tight-junction permeability during nutrient absorption across the intestinal epithelium. Annu Rev Nutr. 1995;15:35–55.
Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Phys Cell Phys. 2004;286:C1213–28.
Ozden O, Black BL, Ashwell CM, Tipsmark CK, Borski RJ, Grubb BJ. Developmental profile of Claudin-3,-5, and-16 proteins in the epithelium of Chick intestine. Anat Rec. 2010;293:1175–83.
Kawasaki K, Hayashi Y, Nishida Y, Miki A, Itoh H. Developmental expression of the tight junction protein, occludin, in the gastrointestinal tract of the chick embryo. Histochem Cell Biol. 1998;109:19–24.
Simard A, Di Pietro E, Ryan AK. Gene expression pattern of Claudin-1 during chick embryogenesis. Gene Expr Patterns. 2005;5:553–60.
Osselaere A, Santos R, Hautekiet V, De Backer P, Chiers K, Ducatelle R, et al. Deoxynivalenol impairs hepatic and intestinal gene expression of selected oxidative stress, tight junction and inflammation proteins in broiler chickens, but addition of an adsorbing agent shifts the effects to the distal parts of the small intestine. PLoS One. 2013;8:e69014.
Du EC, Wang WW, Gan LP, Li Z, Guo SS, Guo YM. Effects of thymol and carvacrol supplementation on intestinal integrity and immune responses of broiler chickens challenged with Clostridium Perfringens. J Anim Sci Biotechnol. 2016;7:19.
McReynolds JL, Byrd JA, Anderson RC, Moore RW, Edrington TS, Genovese KJ, et al. Evaluation of immunosuppressants and dietary mechanisms in an experimental disease model for necrotic enteritis. Poult Sci. 2004;83:1948–52.
Sun Q, Liu D, Guo S, Chen Y, Guo Y. Effects of dietary essential oil and enzyme supplementation on growth performance and gut health of broilers challenged by Clostridium perfringens. Anim Feed Sci Technol. 2015;207:234–44.
Schoster A, Kokotovic B, Permin A, Pedersen PD, Dal Bello F, Guardabassi L. In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe. 2013;20:36–41.
Gerard P, Brezillon C, Quere F, Salmon A, Rabot S. Characterization of Cecal microbiota and response to an orally administered Lactobacillus probiotic strain in the broiler chicken. J Mol Microbiol Biotechnol. 2008;14:115–22.
La Ragione RM, Narbad A, Gasson MJ, Woodward MJ. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett Appl Microbiol. 2004;38:197–205.
Fukata T, Hadate Y, Baba E, Arakawa A. Influence of bacteria on Clostridium-perfringens infections in young chickens. Avian Dis. 1991;35:224–7.
Park SS, Lillehoj HS, Allen PC, Park DW, FitzCoy S, Bautista DA, et al. Immunopathology and cytokine responses in broiler chickens Coinfected with Eimeria maxima and Clostridium perfringens with the use of an animal model of necrotic enteritis. Avian Dis. 2008;52:14–22.
Collier CT, Hofacre CL, Payne AM, Anderson DB, Kaiser P, Mackie RI, et al. Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Vet Immunol Immunopathol. 2008;122:104–15.
Hong YH, Song W, Lee SH, Lillehoj HS. Differential gene expression profiles of beta-defensins in the crop, intestine, and spleen using a necrotic enteritis model in 2 commercial broiler chicken lines. Poult Sci. 2012;91:1081–8.
Lee SH, Lillehoj HS, Jang SI, Lillehoj EP, Min W, Bravo DM. Dietary supplementation of young broiler chickens with capsicum and turmeric oleoresins increases resistance to necrotic enteritis. Bri J Nutr. 2013;110:840–7.
Brisbin JT, Gong J, Parvizi P, Sharif S. Effects of lactobacilli on cytokine expression by chicken spleen and Cecal tonsil cells. Clin Vaccine Immunol. 2010;17:1337–43.
Wang L, Liu C, Chen M, Ya T, Huang W, Gao P, et al. A novel Lactobacillus plantarum strain P-8 activates beneficial immune response of broiler chickens. Int Immunopharmacol. 2015;29:901–7.
Deplancke B, Vidal O, Ganessunker D, Donovan SM, Mackie RI, Gaskins HR. Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am J Clin Nutr. 2002;76:1117–25.
Kim GB, Seo YM, Kim CH, Paik IK. Effect of dietary prebiotic supplementation on the performance, intestinal microflora, and immune response of broilers. Poult Sci. 2011;90:75–82.
Acknowledgements
Not applicable.
Funding
This study was supported by Natural Science Foundation of China (No. 3140130343).
Availability of data and materials
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
ZL designed the study, carried out the experiments and measurements, and drafted the manuscript. WW assisted with the chicken trial and sample collection. DL and YG participated in the study’s design, coordination and paper writing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
The experiment was approved by the China Agricultural University Animal Care and Use Committee.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, Z., Wang, W., Liu, D. et al. Effects of Lactobacillus acidophilus on the growth performance and intestinal health of broilers challenged with Clostridium perfringens. J Animal Sci Biotechnol 9, 25 (2018). https://doi.org/10.1186/s40104-018-0243-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-018-0243-3