Abstract
Background
Intramuscular fat (intramuscular fat, IMF) is one of the important traits of pork quality. How to reasonably improve the intramuscular fat content is the most focus researchers. Some possible regulation of intramuscular fat deposition of candidate genes to cause the attention of people. The objective of this study was to elucidate the relationship between peroxisome proliferator-activated receptor γ (PPARγ) and adipose differentiation-related protein (ADRP) mRNA expression and intramuscular fat (IMF) deposition in the muscle tissue of three breeds of pig: Laiwu (LW), Lulai Black (LL), and Large White (LY).
Results
qPCR analysis of the PPARγ and ADRP genes in the three breeds of pig revealed PPARγ and ADRP mRNA expression profiles of LW > LL > LY and LL > LW > LY, respectively. PPARγ mRNA expression was significantly and positively correlated with IMF deposition (p < 0.05). There were significant correlations between PPARγ and ADRP mRNA expression levels (p < 0.01).
Conclusions
These results suggest correlations between PPARγ and ADRP in fat deposition and regulation in pigs, PPARγ gene may be a main effector of IMF content and play an important role during adipocyte differentiation in pigs, thereby providing new information to further elucidate molecular mechanisms associated with intramuscular fat deposition in Laiwu pigs and provides new data for further molecular studies of mechanisms underlying intramuscular fat deposition in human obesity. The continued elucidation of specific genetic mechanisms between PPARγ and ADRP warrants further studies.
Similar content being viewed by others
Background
Intramuscular fat (IMF) content is an important trait to determine pig meat quality and has been positively correlated with meat tenderness, moisture content, and taste (Wood et al. 2004; Lu et al. 2008; Hausman et al. 2009). Although several candidate genes play only minor roles, together they construct the genetic basis determining fat deposition. Therefore, the search for candidate genes and molecular mechanisms associated with IMF deposition has become an important strategy in IMF research. At present, much research has focused on the identification of candidate genes and underlying mechanisms associated with the regulation of porcine intramuscular fat deposition, with the goal to reasonably improve IMF content.
Peroxisome proliferator-activated receptor γ (PPARγ), as a candidate gene, is a ligand activated transcription factor and a member of the nuclear hormone receptor superfamily (Bispham et al. 2005). Previous research has shown that PPARγ, which is predominantly expressed in adipose tissue and the liver, is a master regulator of adipogenesis and controls adipocyte differentiation, proliferation, and lipid accumulation (He et al. 2013). In addition, PPARγ regulates the expression of genes associated with sugar metabolism and lipid biosynthesis. Recent researches indicate that progranulin (PGRN) is closely related to diabetes and is regarded as a novel adipokine associated with obesity development, affecting adipocyte biology. Also, PPARγ activity can be adjusted by various methods and materials to increase PGRN polypeptide levels (Yang et al. 2013).
Adipose differentiation-related protein (ADRP) is covered in a phospholipid protein lipid droplet surface and can up-regulate fatty acid production (Imamura et al. 2002; Robenek et al. 2006) and promote the formation of lipid droplets for storage of fatty acids (Listenberger et al. 2007).
Previous studies have shown that the PPARγ and ADRP genes play important roles in the regulation of animal lipid deposition. PPARγ activity can be adjusted by various methods and materials, ADRP can up-regulate fatty acid production. Therefore, the objective of the present study was to elucidate the relationship between PPARγ and ADRP mRNA expression and IMF deposition in muscle and to identify the major genes that regulate IMF deposition to improve IMF content in pork.
Laiwu Pig, an excellent type of North China local fatty breed of pig, is known for high meat quality with good tenderness (Zeng et al. 2005), bright color, and higher IMF, while it is the best animal model to study the mechanisms of high intramuscular fat deposition (Spurlock and Gabler 2008; Chen et al. 2013). The Lulai Black pig is a cross of a Laiwu pig with introgression of a Large White pig. As the low intramuscular fat pig animal model, Large Whites (LY) is the best characterized and studied breed of pig (Grzes et al. 2016; Davoli et al. 2015; De Rosa et al. 2013).
This study is the first to investigate PPARγ and ADRP gene expression profiles in Laiwu pigs and to report the effects of PPARγ and ADRP mRNA expression on regulation of fat deposition in three breeds of pig.
Methods
Experimental animals
This work was approved by the Institutional Animal Care and Use Ethics Committee of Shandong Agricultural University and carried out in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, PR China). The experimental cohort consisted of 30 castrated boars composed of equal numbers of three breeds: Laiwu (LW), Lulai Black (LL), and Large Whites (LY). All pigs were housed at Laiwu Breeder Pig Farm Co. Ltd. (Laiwu city, Shandong Province, China) and fed a diet formulated to meet current nutritional requirements (NRC 1998). On the day of slaughter, the mean weight of the pigs was 114 ± 2 kg. Samples were collected from the longissimus dorsi muscle at the last rib and a portion of the muscle tissue. Afterward, the remaining samples were stored at −80 °C for further analysis.
IMF content measurement
IMF was chemically quantified following the ISO, 1443-1973. The method used was direct Soxhlet extraction of fat by a solvent (AOAC 2000).
RNA extraction and cDNA preparation
Total RNA was extracted from the muscle samples using TRIZOL® Reagent (Invitrogen, San Diego, CA, USA) and its integrity was verified in a 1 % agarose gel. Total RNA was reverse transcribed to cDNA using SuperScrit TM III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions (Omi et al. 2005; Cui et al. 2011).
Primer design
Primers for amplification of PPARγ (GenBank_NM214379.1) and ADRP (GenBank_NM214200.1) mRNA were designed using Premier 5.0 software (PREMIER Biosoft, Palo Alto, CA, USA). The primer pair used for amplification of ACTB mRNA is reported elsewhere (AOAC 2000). All primer sequences are listed and described in detail in Table 1, and were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China).
Establishment of a real-time fluorescence quantitative PCR (qPCR) system
SYBR Green real-time PCR amplification was conducted using an Mx3000PTM System (Stratagene Corporation, Santa Clara, CA, USA).The optimal reaction system and conditions were determined by experimentation. The continuously expressed gene, ACTB, was used as an endogenous reference for determination of targeted mRNA profiles. Each qPCR amplification was performed in a 15-μL reaction volume consisting of 7.5 μL of SYBR® Permix Ex Taq™ (TaKaRa, Japan) spiked with 0.3 μL of ROX Reference Dye II (TaKaRa), cDNA 5 μL of PPARγ and ADRP, forward and reverse primers, and 0.3 μL of PPARγ (10 μmol/L) and ADRP (10 μmol/L), respectively, and supplemented with water to a final volume of 15 μL. The following amplification conditions were used: one cycle of 15 s at 95 °C, followed by 40 cycles of 10 s at 95 °C, 20 s at 58 °C, and a final extension for 15 s at 72 °C. The ACTB gene was used as an internal control in the reaction system and amplified in triplicate in accordance with the conditions described above.
Statistical analysis
The results were analyzed using Mx3000P quantitative analysis software and relative quantitative results were calculated using the adjusted \(2^{{ - {\Delta \Delta }{\text{C}}_{\text{T}} }}\) method (Erkens et al. 2006; Livak and Schmittgen 2001). Differential gene expression in muscle tissues collected from the three breeds was subjected to bivariate correlation analysis with repeated measures. Correlation analysis between PPARγ and ADRP mRNA expression levels and meat quality were calculated using the PROC CORR procedure included with the SAS ver. 8.1 software package (SAS Institute, Cary, NC, USA). The results are presented as means ± the standard error of the mean. Probability (p) values of <0.05 and <0.01 were considered significant and extreme differences, respectively.
Results
IMF content measurement
IMF content was chemically quantified in a total of 30 pigs in accordance with the ISO 1443-1973 protocol with direct Soxhlet extraction of fat by a solvent. As shown in Table 2, the average IMF content of LW was significantly higher than that of LL and LY, indicating significant differences among breeds (p < 0.05).
PPARγ and ADRP mRNA expression level
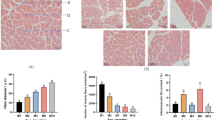
As shown in Fig. 1, there were significant differences in PPARγ mRNA expression levels among breeds (p < 0.05), with highest values in LW followed by LL and LY. Also, there was significant difference in ADRP mRNA expression levels among breeds, with highest values in LL, followed by LY and LW (p > 0.05) (Fig. 2).
PPARγ and ADRP mRNA expression correlated with IMF content
The results of correlation analysis between PPARγ and ADRP mRNA expression with IMF content are shown in Table 3. PPARγ mRNA expression in muscle was positively correlated with IMF (p < 0.05) in all three breeds of pig. ADRP mRNA expression in muscle was not correlated with IMF. With regard to IMF, LW was significantly thicker than LY, there were significant differences between LW and LL. These results imply that the ADRP gene is not influenced by a variety of factors, while PPARγ expression was significantly correlated with IMF content (p < 0.01).
Discussion
PPARγ and ADRP mRNA expression
The results of this study showed that mRNA levels of PPARγ and ADRP in the muscle tissues of the three breeds of examined pigs occurred in the following orders, LW > LL > LY and LL > LW > LY, respectively. PPARγ belongs to a nuclear receptor superfamily of transcription factors and plays a critical role in adipocyte differentiation and fat deposition in mammals (Farmer 2006; Zeng et al. 2012).
PPARγ mRNA expression was significantly correlated with IMF in muscle in each of the three breeds of pig, suggesting some type of relationship between the gamma component, which may be related to the fat deposition traits associated with PPAR. PPARγ is mainly involved in the induction of differentiation of adipose cells as well as activation of the expression of phosphoenolpyruvate and other fatty tissue-specific genes (Pan et al. 2014). Meanwhile, Ma et al. (2015) found that the PPARγ-BsrI loci can be used as a candidate gene loci of pork quality traits.
PPARγ mRNA expression is correlated with IMF content
PPARγ mRNA expression in muscle was significantly and positively correlated with IMF (p < 0.05) in the three breeds of pig, further confirming that PPARγ plays a certain role in fat traits. Farmer (2006) and Kim et al. (2005) pointed out that the gamma component is extremely important in anomalies in the process of differentiation of preadipocytes and adipocyte maturation associated with the complex PPAR transcription factor, and can directly or indirectly regulate other transcription factors to induce activation of fat cells. The results of a study of Rongchang piglets showed that increased gene expression of PPARγ in adipose tissues may regulate fat deposition throughout the whole body (Bildirici et al. 2003). Another study reported that miRNA-130b suppressed fat deposition by inhibiting PPARγ expression and demonstrated a significant ability to down-regulate PPARγ expression, which was associated with reduced adipogenesis and lipogenesis in primary cultured porcine adipocytes (Rosen and Spiegelman 2001; Taniguchi et al. 2014).
Together, these results suggest that the PPARγ gene may be a main effector of IMF content and play an important role during adipocyte differentiation in pigs, thereby providing new information to further elucidate molecular mechanisms associated with intramuscular fat deposition in Laiwu pigs.
By confirming a functional pig muscle-specific promoter, we could drive fat-related gene overexpression in skeletal muscle and improve IMF content via the development of transgenic pigs.
ADRP mRNA expression is not correlated with IMF content
There was no significant correlation between ADRP mRNA expression in muscle and IMF (p > 0.05) and there were significant correlations with PPARγ and ADRP mRNA expression levels (p < 0.01). ADRP plays an important role in regulating lipid storage in various cells. Kim et al. (2005) investigated ADRP as a candidate gene of intramuscular fat deposition and marbling traits in pigs. Results showed that, in the aspect of human studies (Yuan et al. 2008), ADRP gene expression was mediated by PPARγ. Also, PPARγ is a very important mediating factor in the resistance to insulin. Research of human trophoblasts showed that ADRP expression was regulated by PPARγ/Retinoid X receptor.
Conclusions
These results suggest correlations between PPARγ and ADRP in fat deposition and regulation in pigs. PPARγ gene may be a main effector of IMF content and play an important role during adipocyte differentiation in pigs, thereby providing new information to further elucidate molecular mechanisms associated with intramuscular fat deposition in Laiwu pigs and provides new data for further molecular studies of mechanisms underlying intramuscular fat deposition in human obesity. The continued elucidation of specific genetic mechanisms between PPARγ and ADRP warrants further studies.
Abbreviations
- PPARγ:
-
peroxisome proliferator-activated receptor γ
- ADRP:
-
adipose differentiation-related protein
- IMF:
-
intramuscular fat
- LW:
-
Laiwu pig
- LL:
-
Lulai Black pig
- LY:
-
Large White
- PGRN:
-
progranulin
References
AOAC (2000) Official methods of analysis of the association of analytical chemists, 17th edn. AOAC Int, Washington
Bildirici I, Roh CR, Schaiff WT, Lewkowski BM, Nelson DM, Sadovsky Y (2003) The lipid droplet-associated protein Adipophilin is expressed in human trophoblasts and is regulated by peroxisomal proliferator-activated receptor-/retinoid X receptor. J Clin Endocrinol Metab 88:6056–6062
Bispham J, Gardner DS, Gnanalingham MG, Stephenson T, Symonds ME, Budge H (2005) Maternal nutritional programming of fetal adipose tissue development: differential effects on messenger ribonucleic acid abundance for uncoupling proteins and peroxisome proliferator-activated and prolactin receptors. Endocrinology 146:3943–3949
Chen QM, Zeng YQ, Wang H, Yang L, Yang Y, Zhu HL, Shi Y, Chen W, Hu YX (2013) Molecular characterization and expression analysis of NDUFS4 gene in m. longissimus dorsi of Laiwu pig (Sus scrofa). Mol Biol Rep 40:1599–1608
Cui JX, Zeng YQ, Wang H, Chen W, Du JF, Chen QM, Hu YX, Yang L (2011) The effects of DGAT1 and DGAT2 mRNA expression on fat deposition in fatty and lean breeds of pig. Livest Sci 140:292–296
Davoli R, Luise D, Mingazzini V, Zambonelli P, Braglia S, Serra A, Russo V (2015) Genome-wide study on intramuscular fat in Italian Large White pig breed using the PorcineSNP60 BeadChip. J Anim Breed Genet 133:277–282
De Rosa A, Monaco ML, Nigro E, Scudiero O, D′Andrea M, Pilla F, Oriani G, Daniele A (2013) Tissue-specific downregulation of the adiponectin “system”: possible implications for fat accumulation tendency in the pig. Domest Anim Endocrinol 44:131–138
Erkens T, Mario VP, Jo Vandesompele, Goossens K, Zeveren AV, Peelman LJ (2006) Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol 6:41–48
Farmer SR (2006) Transcriptional control of adipocyte formation. Cell Metab 4:263–273
Grzes Maria, Sadkowski Slawomir, Rzewuska Katarzyna, Szydlowski Maciej, Switonski Marek (2016) Pig fatness in relation to FASN and INSIG2 genes polymorphism and their transcript level. Mol Biol Rep 43:381–389
Hausman GJ, Dodson MV, Ajuwon K, Hausman GJ, Dodson MV, Ajuwon K, Azain M, Barnes KM, Guan LL, Jiang Z, Poulos SP, Sainz RD, Smith S, Spurlock M, Novakofski J, Fernyhough ME, Bergen WG (2009) Board sponsored invited review: the biology and regulation of preadipocytes and adipocytes in meat animals. J Anim Sci 87:1218–1246
He K, Wang QS, Wang Z, Pan YC (2013) Association study between gene polymorphisms in PPAR signaling pathway and porcine meat quality traits. Mamm Genome 24:322–331
Imamura M, Inoguchi T, Ikuyama S, Taniquchi S, Kobayashi K, Nakashima N, Nawata H (2002) ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am J Physiol Endocrinol 283:775–783
Kim TH, Choi BH, Chang GW, Lee KT, Lee HY, Lee JH, Kim KS, Park CK, Moran C (2005) Molecular characterization and chromosomal mapping of porcine adipose differentiation-related protein (ADRP). J Anim Breed Genet 122:240–246
Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA (2007) Adipocyte differentiation-related protein reduces lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res 48:2751–2761
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C (T)) method. Methods 25:402–408
Lu P, Li DF, Yin JD, Zhang LY, Wang ZY (2008) Flavour differences of cooked longissimus muscle from Chinese indigenous pig breeds and hybrid pig breed (Duroc × Landrace × Large White). Food Chem 107:1529–1537
Ma JJ, Chai J, Shang YY, Li YJ, Chen R, Jia J, Jiang S, Peng J (2015) Swine PPAR-γ expression upregulated in skeletal muscle of transgenic mice via the swine Myozenin-1 gene promoter. Transgenic Res 24:409–420
NRC (1998) Nutrient requirements of swine, 10th edn. Nutrient Requirements of Domestic Animals, Washington
Omi T, Brenig B, Spilar Kramer S, Iwamoto S, Stranzinger G, Neuenschwander S (2005) Identification and characterization of novel peroxisome proliferator-activated receptor-gamma (PPAR-gamma) transcriptional variants in pig and human. J Anim Breed Genet 122:45–53
Pan S, Yang X, Jia Y, Li R, Zhao R (2014) Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-g expression. J Cell Physiol 229:631–639
Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ (2006) Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci 119:4215–4224
Rosen ED, Spiegelman BM (2001) PPAR gamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem 276:37731–37734
Spurlock ME, Gabler NK (2008) The development of porcine models of obesity and the metabolic syndrome. J Nutr 138:397–402
Taniguchi M, Nakajima I, Chikuni K, Kojima M, Awata T, Mikawa S (2014) MicroRNA-33b downregulates the differentiation and development of porcine preadipocytes. Mol Biol Rep 41:1081–1090
Wood JD, Nute GR, Richardson RI, Whittington FM, Southwood O, Plastow G, Mansbridge R, Costa N, Chang KC (2004) Effects of breed, diet and muscle on fat deposition and eating quality pigs. Meat Sci 67:651–667
Yang H, Cheng J, Song Z, Li X, Zhang Z, Mai Y, Pang W, Shi X, Yang G (2013) The anti-adipogenic effect of PGRN on porcine preadipocytes involves ERK1, 2 mediated PPARγ phosphorylation. Mol Biol Rep 40:6863–6872
Yuan JS, Wang D, Sewart CN (2008) Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol J 3:112–123
Zeng YQ, Wang GL, Wang CF, Wei SD, Wu Y, Wang LY (2005) Genetic variation of H-FABP gene and association with intramuscular fat content in Laiwu Black and four western pig breeds. Asian-Aust J Anim Sci 18:13–16
Zeng Z, Yu B, Mao X, Chen D (2012) Effects of dietary digestible energy concentration on growth, meat quality, and PPARγ gene expression in muscle and adipose tissues of Rongchang piglets. Meat Sci 90:66–70
Authors’ contributions
JXC conceived and designed the experiments. WC, TX analyzed the data and JL, YQZ edited it. JXC wrote this manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported financially by National Natural Science Foundation of China (Grant No. 31401055), Shandong Province Agricultural Animal Breeding Project of China (Grant No. 2013LZ02-015), Shandong Province Modern Pig Technology and Industry System Project (Grant No. SDAIT-06-022-03), Shandong Province Weifang city science and Technology Bureau Project of China (Grant No. 201301153). Shandong University of science and technology project (Grant No. J16LE58).
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This work was approved by the Institutional Animal Care and Use Ethics Committee of Shandong Agricultural University and carried out in accordance with the ‘‘Guidelines for Experimental Animals’’ of the Ministry of Science and Technology (Beijing, PR China).
Author information
Authors and Affiliations
Corresponding author
Additional information
Jingxiang Cui and Wei Chen contributed equally to this work
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Cui, J., Chen, W., Liu, J. et al. Study on quantitative expression of PPARγ and ADRP in muscle and its association with intramuscular fat deposition of pig. SpringerPlus 5, 1501 (2016). https://doi.org/10.1186/s40064-016-3187-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-3187-0