Abstract
Background
Parkinson’s disease (PD) is characterized by a chronic loss of dopaminergic neurons and the presence of proteinaceous inclusions (Lewy bodies) within some remaining neurons in the substantia nigra. Recently, astroglial inclusion body has also been found in some neurodegenerative diseases including PD. However, the underlying molecular mechanisms of how astroglial protein aggregation forms remain largely unknown. Here, we investigated the contribution of αB-crystallin (CRYAB), a small heat shock protein, in α-synuclein inclusion formation in astrocytes.
Methods
Small interfering RNA (siRNA)-mediated CRYAB (siCRYAB) knockdown or CRYAB overexpression was performed to investigate the impact of CRYAB on the autophagy in human glioblastoma cell line U251 cells. Co-immunoprecipitation (co-IP) and immunoblotting were used to dissect the interaction among multiple proteins. The clearance of α-synuclein in vitro was evaluated by immunocytochemistry. CRYAB transgenic mice and transgenic mice overexpressing A30P mutant form of human α-synuclein were used to examine the influence of CRYAB to α-synuclein accumulation in vivo.
Results
We found that knockdown of CRYAB in U251 cells or primary cultured astrocytes resulted in a marked augmentation of autophagy activity. In contrast, exogenous CRYAB disrupted the assembly of the BAG3-HSPB8-HSC70 complex via binding with BAG3, thereby suppressing the autophagy activity. Furthermore, CRYAB-regulated autophagy has relevance to PD pathogenesis. Knockdown of CRYAB remarkably promoted cytoplasmic clearance of α-synuclein preformed fibrils (PFFs). Conversely, selective overexpression of CRYAB in astrocytes markedly suppressed autophagy leading to the accumulation of α-synuclein aggregates in the brain of transgenic mice expressing human α-synuclein A30P mutant.
Conclusions
This study reveals a novel function for CRYAB as a natural inhibitor of astrocytic autophagy and shows that knockdown of CYRAB may provide a therapeutic target against proteinopathies such as synucleinopathies.
Similar content being viewed by others
Background
Parkinson’s disease (PD) is the most common movement disorder. It is characterized by chronic degeneration of nigral dopaminergic (DA) neurons. There is currently no cure for this devastating neurodegenerative disorder. This is largely due to a limited understanding of its pathogenesis. Over the decades, several hypotheses about PD pathogenesis have been proposed, including mitochondria dysfunction, oxidative stress, abnormal protein aggregation, protein kinase dysfunction, unbalanced calcium homeostasis and neuroinflammation [1]. Abnormal protein aggregation in the remaining nigral DA neurons, known as a Lewy body, is one of the most prominent pathological features in the Parkinsonian brain, in addition to the chronic loss of DA neurons. Over the past few decades, numerous studies have mainly been focused on analyzing protein accumulation and degradation in neuronal cells. This focus is largely attributable to the fact that abnormal aggregates containing disease-causing protein in neuronal cells are the most prominent pathological changes when visualizing the protein aggregates using α-synuclein antibody.
However, accumulating evidence suggests the presence of glial inclusion in PD brain. For example, Braak et al. found the presence of α-synuclein-immunoreactive inclusions in astrocytes in prosencephalic regions [2]. It is hypothesized that astrocytic protein aggregates, such as α-synuclein, are taken up from neurons to be degraded in the lysosome [3, 4]. At advanced disease stages, protein aggregates are accumulated in astrocytes. Indeed, post-mortem brain analysis showed that α-synuclein-immunoreactive inclusions appear frequently in astrocytes as well as neurons in the PD brain [2, 5,6,7,8]. Accumulated protein aggregates are likely to affect astrocyte functions, thereby contributing to neurodegeneration. This notion is supported by several animal studies. For instance, the accumulation of α-synuclein induces astrocytic mitochondrial impairments [4]. Moreover, selective overexpression of an α-synuclein mutant in astrocytes leads to degenerative movement disorder in mice, accompanied by astrogliosis [9]. These data indicate that PD pathogenesis is much more complex than we previously anticipated and glial inclusion may impact on glial function during neurodegeneration thereby contribute to disease progression. It is important to investigate the question of how the glial inclusion is formed during neurodegeneration, a question which has been overlooked in this field for years.
Recent meta-analysis of genome-wide association studies (GWASs) revealed a set of novel risk loci involved in PD susceptibility, in addition to those that have been previously reported at genome-wide significance levels. These data point to a key role for autophagy and lysosomal biology in PD [10]. Interestingly, among the PD risk loci identified, some of the candidate genes, such as Bcl-2-associated athanogene 3 (BAG3), a member of the BAG family of proteins, and GTP cyclohydrolase I (GCH1), are known to be expressed in both astrocytes and neurons. The expression levels of these genes in astrocytes were preferentially upregulated in the Parkinsonian brain along with protein aggregation [11] or in response to other pathological stimuli [12, 13], implying that these candidate genes play a role in the glial pathology of PD.
Autophagy is considered as a major system targeting long-lived proteins and dysfunctional organelles for lysosomal degradation. Impaired autophagy activity and significantly increased protein aggregates are found in α-synucleinopathies as well as in tauopathies and amyotrophic lateral sclerosis (ALS) [14,15,16,17,18]. Moreover, a previous study has shown that a variety of susceptible genes related to degenerative diseases are associated with autophagy-lysosome regulation [19, 20]. Indeed, BAG3 and HSPB8 whose expression was upregulated specifically in astrocytes in the PD brain [11], have been shown to be able to promote autophagy, indicating that alterd autophagy process in astrocytes is involved in the failure of glial inclusion clearance in PD and may thus contribute to disease onset/progression in neurodegeneration. The persistent existence of glial inclusions in the PD brain raises a question as to whether a yet-unidentified mechanism plays a role in this process, despite the upregulation of BAG3/HSPB8-induced autophagy.
It has been shown that αB-crystallin (CRYAB), a small heat shock protein (sHSP), is mainly expressed in astrocytes and oligodendrocytes in the central nervous system [21]. CRYAB is implicated in various protein aggregation-related neurodegenerative diseases such as Alzheimer’s disease (AD), PD, ALS, tauopathies, Alexander’s disease, and prion disorders [2, 21,22,23,24,25,26,27,28]. For example, CRYAB and tau are often co-localized in glia with tau pathology [22]. The relationship between increased glial CRYAB expression and abnormal protein accumulation remains unclear. Previously, we found that expression of CRYAB was steadily increased in the ventral midbrain and striatum of an adult mouse brain with aging [29]. Moreover, we demonstrated that the presence of abnormal CRYAB-immunoreactive inclusions in astrocyte-like glial cells in the substantia nigra (SN) of the PD brain is a remarkable pathological feature of the gliosis in PD [21]. We hypothesized therefore that CRYAB may play a role in glial inclusion formation during brain aging and contribute to PD pathogenesis.
In the present study, we tested this hypothesis by investigating the effect of CRYAB on astrocytic autophagy flux in vitro and on the degradation of α-synuclein mutant protein in vivo. We showed that CRYAB functions as a potent inhibitor of astrocytic autophagy induction through interacting with BAG3, which is a key mediator for macroautophagy (hereafter referred to autophagy) induction. By inhibiting the selective process of autophagy, elevated levels of CRYAB leads to the accumulation of aggregated proteins in astrocytes that exacerbates the pathological process of neurodegenerative diseases such as PD.
Methods
Animals
Adult or neonatal C57BL/6 mice were from Shanghai Laboratory Animal Centre, Chinese Academy of Sciences. CRYAB transgenic mice (FVB-Tg(GFAP-CRYAB)141.6Mes/J) and SNCAA30P (B6.Cg-Tg(THY1-SNCA*A30P)TS2Sud/J) transgenic mice were purchased from the Jackson Laboratory (USA). CRYAB Tg mice in a C57BL/6 (inbred) genetic background generated by 10 backcrosses were used in the entire study. They were maintained on a 12 h light/dark cycle at 23 °C with food and water available ad libitum. All procedures performed were approved by the Institutional Animal Care and Use Committee and were in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Cell culture and transfection
U251 human glioblastoma cell line (Cell Bank, Chinese Academy of Sciences) were maintained in DMEM with 10% fetal bovine serum (FBS) and 100 mM glutamine at 37 °C under 5% CO2. Transient transfection was performed using Lipofectamine 3000 (Invitrogen, L3000–008) according to the manufacturer’s instructions. Knocking down CRYAB was performed as described previously [30]. siRNA duplexes were used (GenePharma, Shanghai, China) and their sequences were as follows: sense: r (GGC CCA AAU UAU CAA GCU A) dTdT; antisense: r (UAG CUU GAU AAU UUG GGC C) dTdG; non-silencing control siRNA: sense: r (UUC UCC GAA CGU GUC ACG U) dTdT; and antisense: r (ACG UGA CAC GUU CGGAGAA) dTdT. At 72 h after transfection, cells were harvested for further analysis.
Plasmids, reagents, and antibodies
The cDNA of BAG3 and CRYAB were amplified using RNA extracted from HEK293 cell, followed by RT-PCR and subcloned into pLenti vector (OBIO, H139).
H2O2 (SinoPharm Chemical Reagent Co., Ltd., China. Cat. #7722-84-1). Rapamycin (Sangon Biotech, China. Cat. #A606203) and chloroquine (Sangon Biotech. Cat. #A506569) were used. The following antibodies were used: anti-LC3 (MBL, Cat. #PM036), anti-β-actin (Sigma, Cat. #A5441), anti-RPS6KB/p70S6 (Cell Signaling, Cat. #2708), anti-phospho-RPS6KB/p70S6 (Cell Signaling, Cat. #9206), anti-mTOR (Cell Signaling, Cat. #2972), anti-phospho-mTOR (Sigma, Cat. #SAB4504043), anti-SQSTM1/p62 (MBL, Cat. #PM045), anti-GAPDH (SAB, Cat. #1336), anti-CRYAB (Santa Cruz Biotech, Cat. #sc-137,143), anti-HSPB8 (Abcam, Cat. #ab4149), anti-HSC70 (Santa Cruz Biotech, Cat. #sc-7298), anti-BAG3 (Abcam, Cat. #ab47124), anti-ubiquitin (DAKO, Cat. #z0458), anti-α-synuclein (Cell Signaling, Cat. #2628), anti-GFP (Santa Cruz Biotech, Cat. #sc-9996).
Western blot analysis and quantification
Western blot was performed as described previously [31]. Briefly, the proteins from lysed cells were denatured and loaded on sodium dodecyl sulfate polyacrylamide gels. Afterwards, the proteins were transferred to PVDF membranes, blocked in TBST (150 mM NaCl, 10 mM Tris-HCl, pH 7.5 and 0.1% Tween 20) containing 5% (w/v) skim milk, and incubated with the corresponding primary and secondary antibodies.
Peroxidase activity was detected with Omni-ECL™ FemtoLight Chemiluminescence Kit (EpiZyme, Cat. #SQ201) and visualized and digitized with ImageQuant (LAS-4000, FujiFilm, Japan). Optical densities of bands were analyzed by using Image J. Protein levels, quantified by computer analysis as the ratio between each immunoreactive band and the levels of β-actin were expressed as a percentage of vehicle-treated control.
Co-immunoprecipitation analysis
To analyze protein interactions in vitro, approximately 5 × 106 U251 cells were harvested after 48 h transfection and lysed in 0.5 ml of RIPA buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). Pre-cleared lysates were subjected to immunoprecipitation with indicated antibodies directed against the epitope tag as described previously. Precipitated proteins were eluted from the beads by boiling in SDS sample buffer, and separated by SDS-PAGE. Immunoblot assays were performed using the indicated antibodies. The following primary antibodies were used: anti-Flag Affinity gel (Genomics Technology, Cat. #SG4510–10), anti-c-Myc AC (Santa Cruz Biotech, Cat. #sc-40 AC).
Primary astrocytic culture and plasmid transfection
Astrocytes were prepared from the cerebrocortex of wild-type C57BL/6 mice at P0, as described previously [32]. The neonatal cortex was trypsinized and dissociated. Cells were then plated at 75 cm2 flask (Corning, USA) in DMEM containing 10% FBS. Culture media were changed 24 h later and subsequently twice a week. Cultured cells were shaken to remove the top layer of cells sitting over the astroglial monolayer to yield mainly type I astrocytes with a flat morphology between day 5 and 7 in vitro. Before experimental treatments, astrocytic cultures were passaged once. Cells were allowed to reach 90% confluence. Cultures were transfected using lentivirus, which can express CRYAB. The cells transfected with the control lentivirus were included as controls in all experiments.
Immunostaining and confocal microscopy
For immunostaining, cells were fixed in 4% paraformaldehyde followed by 0.1% Triton-X100 in PBS (pH 7.4). After washing twice with PBS, they were incubated in FBS (PBS, pH 7.4, containing 5% FBS) to block nonspecific sites of antibody adsorption. Then, the cells were incubated with corresponding primary antibody followed by incubation of secondary antibody conjugated with either Alex488 or Alex555. The same sample were then incubated with another corresponding primary and secondary antibodies. Each sample was incubated with Hoechst to show the nucleus. The cells were imaged with invert confocal microscope (FV1000; Olympus).
For quantification of the number of autophagosomes (diameters 0.3–1.0 μm), a total of 50 cells were analyzed using ImageReader software (Fujifilm). Quantification of LC3 and BAG3 fluorescence intensity and analysis of protein co-localization coefficients in 30 cells were performed using ImageReader software (FujiFilm).
Electron microscopy
Electron microscopic analysis of U251 cells was performed using a standard protocol. In brief, cells were fixed in 2.5% glutaraldehyde in 0.1 M PB (pH 7.4), and post-fixed in aqueous 1% OsO4 followed by 3–4% uranyl acetate. After ethanol and acetone dehydration and embedding in resin, ultrathin (70 nm) sections were then stained with 2% uranyl acetate followed by 0.3% lead citrate. Sections were imaged using transmission electron microscope (Nippon Tekno Co Ltd., model JEOL-1230) at 80 kV. For autophagic vacuole quantification, 20 cells, primary magnification × 20,000, were taken with systematic random sampling from each sample. The cytoplasmic volume fraction of autophagic vacuoles was estimated using ImageReader software (FujiFilm).
Extraction of RIPA-soluble and RIPA-insoluble protein fractions of mouse brain
Extraction of RIPA-soluble and RIPA-insoluble protein fractions from mouse brain tissues was described previously [33,34,35]. Previous studies indicated that separation of insoluble fraction was performed on older mice at least 6 months of age [32, 36]. The tissues from one-year-old CRYABTg; SNCAA30P Tg mice were isolated and stored at -80 °C prior to use. Frozen brain tissues were thawed on ice and homogenized in 10 mL/g (v/w) radio-immunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% TritonX-100, 0.1% SDS, 0.5% sodium deoxycholate, 50 mM NaF, 1 mM EDTA, 50 mM Tris, pH 7.5 with phosphatase and protease inhibitor cocktails). The homogenates were centrifuged at 14,000 g at 4 °C for 30 min and the supernatants were collected as the RIPA-soluble fraction. The pellet was washed with RIPA buffer and centrifuged at 14,000 g at 4 °C for 10 min for three times. The resultant pellet was sonicated in 5 mL/g (v/w) urea buffer (8 M urea, 100 mM NaCl, 1 mM EDTA, 50 mM Tris, pH 8.0) as urea-soluble fraction.
Alpha-synuclein PFFs exposure
Exposure of U251 cells to α-synuclein PFFs was performed as described previously [4, 37]. Briefly, U251 cells were exposed to 0.5 μM α-synuclein PFFs (a gift from Drs. Lee, Trojanowski and Luk, USA) for 24 h. After the treatment, cells were washed twice with culture medium to remove α-synuclein PFFs. On day 0 and 3 in vitro after exposure, cells were fixed for further analyses.
Statistical analysis
Statistical analysis was performed using GraphPad software (GraphPad Prism v6.0; GraphPad Software). All the statistical data were presented as mean ± SEM. Statistical significance of the differences was determined using Student t-test. Differences were considered significant at values of P < 0.05.
Results
CRYAB inhibits astrocytic autophagy
To investigate whether CRYAB has an impact on autophagy in astrocytes, we performed small interfering RNA (siRNA)-mediated CRYAB (siCRYAB) knockdown in U251 cells, which is a cell line derived from human glioblastoma. The conversion of LC3-I to LC3-II was markedly enhanced by CRYAB knockdown (Fig. 1a). In contrast, the overexpression of CRYAB in U251 cells remarkably reduced the conversion of LC3-I to LC3-II in U251 cells compared with cells transfected with an empty vector (Fig. 1b). Consistent with these results, the reduced conversion of LC3-I to LC3-II was also observed in primary cultured astrocytes overexpressing CRYAB via lentivirus-mediated transfection compared with the control (Fig. 1c). These results suggest that CRYAB is a potent regulator of autophagy in astrocytes under normal conditions.
CRYAB regulates autophagy process in vitro. a Knockdown of CRYAB enhances the conversion of LC3-I to LC3-II in U251 cells. U251 cells were transfected with CRYAB siRNA (siNC: the siRNA of negative control; siCRYAB: the siRNA of CRYAB) and the cellular LC3 levels were assessed by Western blotting 48 h after transfection. Data are presented as mean ± SEM, n = 6. Unpaired t-test. **P < 0.01. b Overexpression of CRYAB inhibits LC3-II formation in U251 cells. The cellular LC3 levels were assessed by western blotting 48 h after transfection. Data are presented as the mean ± SEM, n = 3. Unpaired t-test. **P < 0.01. c CRYAB overexpression in primary astrocytes inhibits LC3-II formation. Primary cultured astrocytes were transfected with lentivirus vector encoding CRYAB. The cellular LC3 levels were assessed by Western blotting 48 h after transfection. Data are presented as mean ± SEM, n = 3. Paired t-test. *P < 0.05. d U251 cells were transfected with siCRYAB in the presence or absence of H2O2 (1 mM, 6 h). The cellular p62 levels were assessed 48 h after transfection using western blotting. Data are presented as mean ± SEM, n = 3. *P < 0.05. e Knockdown of CRYAB leads to a marked increase in the number of autophagic vacuoles. Autophagic vacuoles in U251 cells transfected with siNC or siCRYAB were examined using electron microscopy. The arrows indicate autophagic vacuoles. Scale bars: 0.5 μm. The graph shows a statistical analysis of cytoplasmic occupancy of autophagic vacuoles in the cells. Data are presented as mean ± SEM. Twenty cells in each group were examined. Unpaired t-test. ***P < 0.001
We then asked whether CRYAB is capable of influencing autophagy in astrocytes under stress conditions. Hydrogen peroxide (H2O2) has been widely used to induce autophagy in vitro [38]. We evaluated the autophagy activity by measuring p62 levels by using immunoblotting. p62 is known as an autophagy substrate which serves as a reporter of autophagy activity. Promotion of autophagy induced by knockdown of CRYAB resulted in a significant reduction (~ 15%) in p62 levels compared with control (Fig. 1d). Exposure to H2O2 of U251 cells in combination with CRYAB knockdown led to a further reduction in p62 levels (~ 45%), as compared to H2O2 treatment alone. These results suggest that CRYAB also inhibits autophagy in U251 cells under pathological conditions.
To validate whether CRYAB knockdown truly induces autophagy, we performed electron microscopy analysis on U251 cells. It was revealed that knockdown of CRYAB induced robust formation of autophagic vesicles, mature autophagic vesicles and autolysosomes with visible cytoplasmic contents. The total cytoplasmic occupancy of the intracellular autophagic vacuoles was doubled in siCRYAB-treated cells compared with the control (Fig. 1e). Taken together, these data indicate that CRYAB is a crucial autophagic regulator in astrocytes.
CRYAB inhibits autophagy in a mTOR-independent manner
Next, we sought to determine the mechanisms by which CRYAB regulates autophagy. It is well known that mTOR is an important modulator of autophagy [39]. To investigate whether the CRYAB-mediated inhibition of autophagy is dependent on the mTOR pathway in astrocytes, we assessed the effect of rapamycin, an inhibitor of mTOR on autophagy, in U251 cells treated with and without siCRYAB. As expected, siCRYAB strongly elevated autophagy levels, as evidenced by significant increases in the number of LC3-positive puncta (Fig. 2a, c) compared with that number in untreated cell. Concomitant treatment with rapamycin further elevated autophagy levels, as shown by the number of LC3-positive puncta (Fig. 2b, d) and the ratio of LC3-II / LC3-I (Fig. 2e). In contrast, CRYAB knockdown did not profoundly alter the levels of phosphorylated mTOR and phosphorylated p70S6K (Fig. 2f). These data indicate that the molecular mechanisms responsible for CRYAB knockdown-mediated promotion of autophagy is distinct from a well-established mechanism found in rapamycin-treated cells. Interestingly, CRYAB overexpression profoundly attenuated rapamycin-induced autophagy (Fig. 2g).
CRYAB inhibits autophagy in a mTOR-independent manner. a-b Knockdown of CRYAB in U251 cells enhances the formation of LC3+ dots. U251 cells were transfected with siCRYAB. Some of these cells were also treated with 1 μM rapamycin for 6 h. The cells were immunostained with CRYAB and LC3 antibodies 48 h after transfection. Scale bars: 5 μm (a) and 10 μm (b). c-d Quantitative data in a and b, respectively. Data are presented as the mean ± SEM, c, n = 50 cells, d, n = 40 cells. Unpaired t-test. *P < 0.05. e Treatment with rapamycin does not alter CRYAB knockdown of CRYAB-induced augmentation in the conversion of LC3-I to LC3-II in U251 cells. U251 were transfected with siCRYAB and stimulated with 1 μM rapamycin for 6 h. At 48 h after transfection, the cellular LC3 levels were assessed by western blot. The graph shows a statistical analysis of the conversion of LC3-I to LC3-II in the cells. Data are presented as mean ± SEM, n = 5. f Immunoblot analysis of the cell lysates from U251 cells transfected with siCRYAB. The graph shows a statistical analysis of the expression levels of phosphorylated mTOR, p70S6K and total mTOR. Data are presented as mean ± SEM, n = 3. *P < 0.05. g Immunoblot analysis of the cell lysates from U251 cells overexpressing CRYAB and treated with rapamycin (1 μM, 6 h). Data are presented as mean ± SEM, n = 3. *P < 0.05. h Knockdown of CRYAB-induced enhancement in autophagy does not affect autophagic degradation and autophagosome-lysosome fusion. Western blotting analysis of the conversion of LC3-I to LC3-II in U251 cells transfected with siCRYAB and stimulated with 10 μM CQ for 6 h. Data are presented as mean ± SEM. n = 3. *P < 0.05
Moreover, we determined whether CRYAB suppresses autophagy through the inhibition of autophagosome-lysosome fusion. We found that in the presence of chloroquine (CQ), an inhibitor for the autophagosome-lysosome fusion, CRYAB knockdown further augmented the accumulation of autophagosomes compared with CQ treatment alone (Fig. 2h), suggesting that CRYAB does not regulate autophagy through the inhibition of autophagolysosome degradation. Taken together, these results suggest that CRYAB may regulate autophagy in the upstream of mTOR pathway.
CRYAB inhibits BAG3-dependent autophagy via disruption of the assembly of chaperon complex BAG3-HSPB8-HSC70
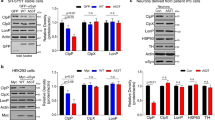
Recent studies have shown that BAG3 potently stimulates macroautophagy [40,41,42,43], an effect that resembles the phenotypes observed in CRYAB knockdown cells (Fig. 1a). Moreover, BAG3 is known to bind to wild-type CRYAB and CRYAB mutant R120G to inhibit CRYAB mutant-induced toxicity by degrading the mutant CRYAB in rat neonatal cardiomyocytes in an autophagy-independent manner in this setting [44]. These findings led us to hypothesize that CRYAB may be functionally related to the BAG3 signalling of autophagy regulation in astrocytes. To test the hypothesis, we first verified the association between BAG3 and CRYAB in astrocytes. As predicted, co-immunoprecipitation (co-IP) assays using cell lysates from U251 cells stably overexpressing CRYAB-Flag revealed that the two proteins bound to each other (Fig. 3a). Immunocytochemical staining showed the co-localization of CRYAB and BAG3 in cultured U251 cells (Fig. 3b).
Overexpression of CRYAB inhibits binding between BAG3 and HSC70 in vitro. a Co-immunoprecipitation (co-IP) reveals that CRYAB interacts with BAG3 in U251 cells. Data are from three independent experiments. b Double immunofluorescence staining. Some CRYAB+ (green) cells (arrowheads indicate examples) express BAG3 (red) in the U251 cell cultures. Right panel is an enlarged view of the box in the left panel. Scale bars: 10 μm. c Co-IP analysis reveals that BAG3 binds to HSC70 and HSPB8. d Co-IP analysis reveals that CRYAB overexpression inhibits binding between BAG3 and HSC70 in U251 cells. Data are from three independent experiments
Moreover, previous studies have shown that BAG3-mediated autophagy is required by its interaction with HSC70 and HSPB8 [42, 43]. It was observed that BAG3 did bind to HSC70 and HSPB8 in U251 cells (Fig. 3c). Thus, we speculated that CRYAB may bind to BAG3 and competitively inhibit the formation of BAG3-HSPB8-HSC70 complex. To test this, we performed BAG3 co-IP assays using U251 cell lines stably overexpressing CRYAB-Flag or empty vector as control. It was found that HSC70, HSPB8 and CRYAB could be detected in BAG3 immunoprecipitates and the levels of HSC70 were decreased in the immunoprecipitates from cells overexpressing CRYAB (Fig. 3d-e). These results suggest that elevation of CRYAB levels can disrupt the assembly of the protein complex BAG3-HSPB8-HSC70.
To functionally characterize the inhibitory effect of CRYAB on BAG3-dependent autophagy, we transfected U251 cells with cDNAs encoding BAG3 and CRYAB concomitantly or separately. Consistent with previous studies, autophagy was markedly increased in BAG3-overexpressing cells compared with the control (Fig. 4a-d). In contrast, BAG3-elevated autophagy was remarkably suppressed by CRYAB overexpression compared with cells transfected with BAG3 alone (Fig. 4a-d), as demonstrated by the number of LC-3-positive puncta (Fig. 4a-b) and the ratio of LC3-II / LC3-I (Fig. 4c-d). These results suggest that CRYAB is a potent inhibitor of BAG3-induced autophagy. Taken together, CRYAB binds to BAG3, preventing the formation of the BAG3-HSPB8-HSC70 complex, thereby inhibiting autophagy in U251 cells.
CRYAB inhibits the BAG3-dependent autophagy in U251 cells. a U251 cells expressing CRYAB stable were transfected with plasmids encoding BAG3-Myc-2A-GFP. The cultures were fixed 48 h after transfection and immunostained with GFP and LC3 antibodies. Scale bars: 10 μm. b Quantification of the number of LC3+ dots per cell shown in (A). Data are presented as mean ± SEM, n = 30 cells. c, d Representative immunoblot shows that BAG3 overexpression elevates the conversion of LC3-II to LC3-I in U251 cells. Cells were transfected with BAG3 plasmids for 48 h. Data are presented as mean ± SEM, n = 5. Unpaired t-test. **P < 0.01
CRYAB inhibits the degradation of aggregated proteins in astrocytes by regulating autophagy
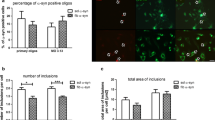
Given that CRYAB is able to regulate autophagy in vitro as shown in the present study, we then investigated whether CRYAB has an impact on the autophagic degradation of α-synuclein in astrocytes. U251 cells were transfected with CRYAB siRNA followed by exposure to 0.4 μM α-synuclein pre-formed fibrils (PFFs) [45] for 24 h. Some cells were maintained for an additional 3 days in the absence of α-synuclein PFFs before fixation (Fig. 5a). Quantifications of α-synuclein immunosignals revealed that CRYAB knockdown accelerated the degradation of α-synuclein on day 3 following incubation with α-synuclein PFFs compared with the control, while no significant alteration of α-synuclein immunosignals was observed on day 0 in siCRYAB-treated cell cultures compared with control (Fig. 5a-e). Moreover, inhibition of autophagosome-lysosome fusion with CQ markedly reduced autophagic degradation of α-synuclein PFFs. This inhibitory effect was not significantly attenuated by CRYAB knockdown (Fig. 5a, f, g). These data suggest that CRYAB is required for the accumulation of α-synuclein in astrocytes through the inhibition of autophagy.
Knockdown of CRYAB in astrocytes promotes α-synuclein degradation. a A schematic shows the timeline of treatment of U251 cells. Cells were transfected with siRNA of CRYAB were exposed to 0.4 μM α-synuclein PFFs for 24 h, washed thoroughly, and cultured for an additional 0 and 3 days in α-synuclein-free medium with 10 μM CQ stimulation before fixation. b-c The intracellular location of the α-synuclein after ingestion in day 0 and day 3 without CQ stimulation was confirmed with confocal imaging. Scale bars: 1 μm. The graph shows a statistical analysis of the mean intensity of the intracellular α-synuclein per field in cells. Data are presented as mean ± SEM, n = 40. d The intracellular location of the α-synuclein after ingestion in day 0 and day 3 without CQ stimulation was confirmed with confocal imaging. The graph shows a statistical analysis of the mean intensity of the total area of the α-synuclein deposits per cell. Data are presented as mean ± SEM, n = 40. e The graph shows a statistical analysis of the mean intensity of the number of α-synuclein deposits per cell. Data are presented as mean ± SEM, n = 40. f, g The intracellular location of the α-synuclein after ingestion on day 3 with CQ stimulation was confirmed with confocal imaging. Scale bars: 1 μm. The graph shows a statistical analysis of the mean intensity of the intracellular α-synuclein per field in cells. Data are presented as mean ± SEM, n = 40. h Western blotting analysis shows that knockdown CRYAB does not affect BAG3 expression levels. Data are expressed as mean ± SEM, n = 3. n.s. no significance. *P > 0.05
To determine whether the impact of CRYAB knockdown on α-synuclein degradation is attributable to enhanced expression of BAG3 which is known to promote autophagy, we assessed the expression levels of BAG3 after CRYAB knockdown in U251 cells. We found that the expression levels of BAG3 were not significantly altered after CRYAB knockdown (Fig. 5h), suggesting that CRYAB knockdown-induced augmentation of α-synuclein aggregate clearance result from elevated BAG3 activity but not its expression.
Astrocytic CRYAB promotes the accumulation of α-synuclein in vivo
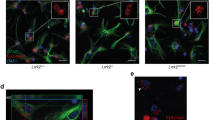
Next, we extended our investigation in vivo to evaluate the impact of CRYAB in astrocytes on the accumulation of α-synuclein using a transgenic mouse model expressing human α-synuclein mutant. We crossed transgenic mice overexpressing the hamster Cryab gene under the control of the human glial fibrillary acidic protein (hGFAP) promoter to target expression preferentially to astrocytes (hereafter referred to as CRYABTg) [46] with transgenic mice overexpressing A30P mutant form of human α-synuclein (SNCAA30P Tg) [47]. Overexpression of CRYAB markedly increased the accumulation of RIPA-insoluble α-synuclein in multiple brain regions, including the ventral midbrain, and striatum of 12-month-old double transgenic mice compared with SNCAA30P Tg mice which served as control (Fig. 6a-b). These results indicate that CRYAB is sufficient for the inhibition of the autophagic degradation of α-synuclein in astrocytes.
Preferential overexpression of CRYAB in astrocytes promotes protein aggregation of a-synuclein in SNCAA30P mutant mouse brain. a Expression levels of RIPA-soluble and RIPA-insoluble protein fractions extracted from the ventral midbrain (VM) in CRYABTg / SNCAA30P Tg+. The graph shows a statistical analysis of the conversion of insoluble to soluble α-synuclein in the tissue. Data are presented as mean ± SEM, n = 5. b Expression levels of RIPA-soluble and RIPA-insoluble protein fractions extracted from the striatum in CRYABTg / SNCAA30P Tg+. The graph shows a statistical analysis of the conversion of insoluble to soluble α-synuclein in the tissue. Data are presented as mean ± SEM, n = 5
Discussion
It has been well recognized that misfolded proteins and abnormal protein aggregation in neuronal cells are the hallmarks of pathology in various neurodegenerative diseases, such as AD, PD and ALS. Interestingly, accumulating evidence has suggested that abnormal protein aggregation also occurs in astrocytes, at least in the advanced stages of the diseases. However, the molecular mechanisms underlying the aberrant accumulation of disease-associated proteins remain largely unknown. In the present study, we demonstrated that CRYAB in astrocytes potently inhibits autophagy and contributes to protein aggregation and neurodegeneration. Mechanistically, CRYAB inhibited the assembly of the functional complex BAG3-HSPB8-HSC70 by binding to BAG3, leading to the deregulation of BAG3-induced autophagy (Fig. 7). These data indicate that the CRYAB-induced suppression of autophagy plays a key role in the aberrant accumulation of α-synuclein in astrocytes, which is a critical pathological event involved in the neurodegeneration of PD.
Model for the role of CRYAB in the clearance of protein aggregates in astrocytes via interaction with BAG3, HSC70 and HSPB8 under both physiological and pathological conditions. During aging and aging-related disease, deregulation of CRYAB in astrocytes may result in a suppression of autophagy leading to the accumulation of α-synuclein proteins and/or organelles
Previous studies suggest that CRYAB does not regulate autophagy in neonatal rat ventricular myocytes [48]. However, we found CRYAB to be required for the regulation of autophagy in astrocytes; as in an adult mouse brain CRYAB is enriched in astrocytes and oligodendrocytes, providing a molecular basis for the CRYAB-dependent inhibition of astrocytic autophagy. These data demonstrate the intriguing specialization of CRYAB function in the brain. CRYAB participates in the regulation of astrocytic autophagy of the brain, representing a mechanistically distinct molecular machinery utilized in the regulation of autophagy in astrocytes.
Numerous studies have shown that sHSPs, including CRYAB, are often considered to be molecular chaperones, that participate in the ubiquitin-proteasome degradation process. sHSPs help hydrolyse misfolded proteins by recognizing and combining with them, to prevent cellular toxicity under certain stress conditions [49, 50]. CRYAB also plays a role in inhibiting inflammation in the process of degenerative diseases [29, 51,52,53]. In the present study, we demonstrated a novel function for CRYAB in astrocytic autophagy regulation. The discrepancy between our findings in the present study and others suggests multifaceted functions of CRYAB. CRYAB plays distinct functions depending on circumstances related to the nature of insults, site of injury and disease staging. We speculate that, under normal conditions or at an early stage of PD, CRYAB functions as a molecular chaperone, participating in the ubiquitin proteasome pathway for misfolded protein degradation. Additionally, it inhibits inflammation to maintain cellular homeostasis under stress conditions. While at the advanced disease stages, despite of the increased levels of CRYAB, CRYAB is not able to prevent the progression of neuroinflammation. Meanwhile, as the protein aggregations become a more severe burden, they are released into the extracellular space and taken up by glial cells, such as astrocytes and microglia [2, 4]. Elevated levels of CRYAB in astrocytes prevent aggregated proteins from degradation by inhibiting autophagy, which in turn accelerates the degeneration of both neuronal and glial cells.
How the function of CRYAB switches is currently unclear. It may be attributable to a particular state of CRYAB determined by its phosphorylation at different sites. This is supported by recent studies showing that CRYAB in different phosphorylation forms plays distinct roles. Levels of CRYAB ser59 are increased significantly under pathological conditions and it plays a role in accelerating inflammation [54]. Therefore, it is necessary to distinguish the different phosphorylation forms of CRYAB in the various context of neurodegenerative diseases.
As shown in Fig. 6, we found that CRYAB derived from astrocytes enhanced the accumulation of insoluble α-synuclein in SNCAA30P Tg mice driven by Thy1 promoter. Given that Thy1 promoter-driven expression of Cre recombinase is largely restricted to neurons [55], it raises the question as to why astrocytic CRYAB promotes α-synuclein accumulation in neuronal cells. Interestingly, a study, using luciferase reporter mice produced by Thy1-Cre mice, showed that Thy1 promoter-driven expression of luciferase reporter gene can be detected in both neurons and astrocytes [56], suggesting that Thy1 promoter is not exclusive for targeting gene expression in neurons, as previously expected. This is supported by a direct evidence showing that in SNCAA30P Tg mice, accumulated α-synuclein could be detected in both astrocytes and neurons on the midbrain and brainstem sections [4]. Thus, an alternative explanation for the results shown in Fig. 6 is that astrocytic CRYAB promotes the accumulation of insoluble α-synuclein in astrocytes of SNCAA30P Tg mice in a cell-autonomous fashion.
We showed in the present study that CRYAB inhibits BAG3-mediated selective autophagy which is an adaptive process to maintain protein homeostasis under stress [57, 58]. The BAG3-HSPB8-HSP70 complex promotes thedegradation of aggregated proteins which is vital for age-related neurodegenerative diseases. During aging, theexpression of BAG3 is induced under oxidative stress and plays a neuroprotective role [41]. Through inhibition of proteasome system, BAG3 can also be upregulated along with aggresome provoking [41]. Through inhibition of proteasome system, BAG3 can also be upregulated along with aggresome provoking [59]. The overexpression of BAG3 in primary cultured neurons significantly promotes Tau degradation In an ALS model, neuronal BAG3 expression is higher in the spinal cords of mutant SOD transgenic mice [43]. A misfolded proteins, such as mutant SOD can also be targeted by BAG3/HSC70, following autophagic degradation [43]. Thus, BAG3-mediated macroautophagy has been considered as a potential therapeutic target for neurodegenerative diseases. Interestingly, during aging or in degenerative diseases, BAG3-HSPB8 expression was more significantly increased in astrocytes than neurons, suggesting that astrocytic autophagy regulation is involved in neurodegeneration and actively responds to extracellular aggregates [11]. Our results showed that the selective overexpression of CRYAB in astrocytes resulted in the heavier accumulation of α-synuclein in aged mice, indicating that suppression of astrocytic autophagy can lead to the aggregation of α- synuclein. Our findings provide new insight into the role of astrocytic autophagy in the degradation of abnormal protein aggregates.
Conclusions
In summary, our study demonstrates that CRYAB is a potent inhibitor of astrocytic autophagy via interacting with BAG3, demonstrating a new signalling pathway that negatively regulates the degradation of α-synuclein aggregates. The present study suggests that the balance between CRYAB-BAG3 and BAG3-HSPB8-HSC70 complexes in astrocytes is an integral components of neuropathology in neurodegeneration. Aberrant increases in the levels of CRYAB inhibit astrocytic autophagy, leading to more severe protein aggregation. The findings that knockdown of astrocytic CRYAB promotes the degradation of aggregated proteins suggest that CRYAB may be a new therapeutic target for future intervention in PD.
Abbreviations
- AD:
-
Alzheimer’s disease
- ALS:
-
Amyotrophic lateral sclerosis
- BAG3:
-
Bcl-2-associated athanogene 3
- co-IP:
-
Co-immunoprecipitation
- CRYAB:
-
αB-crystallin
- DA:
-
Dopaminergic
- PD:
-
Parkinson’s disease
- PFFs:
-
Preformed fibrils
- sHSPs:
-
Small heat shock protein
- siCRYAB:
-
Small interfering RNA (siRNA) targeting CRYAB
References
Maiti P, et al. Current understanding of the molecular mechanisms in Parkinson's disease: targets for potential treatments. Transl Neurodegener. 2017;6:28.
Braak H, et al. Development of alpha-synuclein immunoreactive astrocytes in the forebrain parallels stages of intraneuronal pathology in sporadic Parkinson's disease. Acta Neuropathol. 2007;114(3):231–41.
Lee HJ, et al. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285(12):9262–72.
Lindstrom V, et al. Extensive uptake of alpha-synuclein oligomers in astrocytes results in sustained intracellular deposits and mitochondrial damage. Mol Cell Neurosci. 2017;82:143–56.
Booth HDE, et al. The role of astrocyte dysfunction in Parkinson's disease pathogenesis. Trends Neurosci. 2017;40(6):358–70.
Hishikawa N, et al. Widespread occurrence of argyrophilic glial inclusions in Parkinson's disease. Neuropathol Appl Neurobiol. 2001;27(5):362–72.
Wakabayashi K, et al. NACP/alpha-synuclein-positive filamentous inclusions in astrocytes and oligodendrocytes of Parkinson's disease brains. Acta Neuropathol. 2000;99(1):14–20.
Braidy N, et al. Uptake and mitochondrial dysfunction of alpha-synuclein in human astrocytes, cortical neurons and fibroblasts. Transl Neurodegener. 2013;2(1):20.
Gu XL, et al. Astrocytic expression of Parkinson's disease-related A53T alpha-synuclein causes neurodegeneration in mice. Mol Brain. 2010;3:12.
Chang D, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49(10):1511–6.
Seidel K, et al. The HSPB8-BAG3 chaperone complex is upregulated in astrocytes in the human brain affected by protein aggregation diseases. Neuropath Appl Neuro. 2012;38(1):39–53.
Foster JA, et al. GTP cyclohydrolase I induction in striatal astrocytes following intrastriatal kainic acid lesion. J Chem Neuroanat. 2002;24(3):173–9.
Chiarini A, et al. Proteomic analysis of GTP cyclohydrolase 1 multiprotein complexes in cultured normal adult human astrocytes under both basal and cytokine-activated conditions. Proteomics. 2009;9(7):1850–60.
Lee JH, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141(7):1146–58.
Winslow AR, Rubinsztein DC. The Parkinson disease protein alpha-synuclein inhibits autophagy. Autophagy. 2011;7(4):429–31.
Corrochano S, et al. Alpha-Synuclein levels affect autophagosome numbers in vivo and modulate Huntington disease pathology. Autophagy. 2012;8(3):431–2.
Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–4.
Wu D, et al. Loss of VAPB regulates autophagy in a Beclin 1-dependent manner. Neurosci Bull. 2018;34(6):1037–46.
Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–97.
Fang Z, et al. Neuroprotective Autophagic flux induced by hyperbaric oxygen preconditioning is mediated by cystatin C. Neurosci Bull. 2018; [Epub ahead of print].
Liu Y, et al. Upregulation of alphaB-crystallin expression in the substantia nigra of patients with Parkinson's disease. Neurobiol Aging. 2015;36(4):1686–91.
Dabir DV, et al. Expression of the small heat-shock protein alphaB-crystallin in tauopathies with glial pathology. Am J Pathol. 2004;164(1):155–66.
Iwaki T, et al. Accumulation of alpha B-crystallin in central nervous system glia and neurons in pathologic conditions. Am J Pathol. 1992;140(2):345–56.
Head MW, et al. Overexpression and abnormal modification of the stress proteins alpha B-crystallin and HSP27 in Alexander disease. Am J Pathol. 1993;143(6):1743–53.
Iwaki T, et al. Alpha B-crystallin is expressed in non-lenticular tissues and accumulates in Alexander’s disease brain. Cell. 1989;57(1):71–8.
Renkawek K, et al. Dementia, gliosis and expression of the small heat shock proteins hsp27 and alpha B-crystallin in Parkinson’s disease. Neuroreport. 1999;10(11):2273–6.
Shinohara H, et al. Alpha B crystallin and HSP28 are enhanced in the cerebral cortex of patients with Alzheimer's disease. J Neurol Sci. 1993;119(2):203–8.
Wang K, et al. Abnormally upregulated alphaB-crystallin was highly coincidental with the astrogliosis in the brains of scrapie-infected hamsters and human patients with prion diseases. J Mol Neurosci. 2013;51(3):734–48.
Shao W, et al. Suppression of neuroinflammation by astrocytic dopamine D2 receptors via alphaB-crystallin. Nature. 2013;494(7435):90–4.
Dimberg A, et al. alphaB-crystallin promotes tumor angiogenesis by increasing vascular survival during tube morphogenesis. Blood. 2008;111(4):2015–23.
Li A, et al. Apomorphine-induced activation of dopamine receptors modulates FGF-2 expression in astrocytic cultures and promotes survival of dopaminergic neurons. FASEB J. 2006;20(8):1263–5.
Ho A, et al. Deletion of mint proteins decreases amyloid production in transgenic mouse models of Alzheimer's disease. J Neurosci. 2008;28(53):14392–400.
Bandopadhyay R. Sequential extraction of soluble and insoluble alpha-Synuclein from parkinsonian brains. J Vis Exp. 2016;107:53415.
Neumann M, et al. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117(2):137–49.
Walker AK, et al. An insoluble frontotemporal lobar degeneration-associated TDP-43 C-terminal fragment causes neurodegeneration and hippocampus pathology in transgenic mice. Hum Mol Genet. 2015;24(25):7241–54.
Chandra S, et al. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell. 2005;123(3):383–96.
Rostami J, et al. Human astrocytes transfer aggregated alpha-Synuclein via tunneling nanotubes. J Neurosci. 2017;37(49):11835–53.
Vasko R, Goligorsky MS. Dysfunctional lysosomal autophagy leads to peroxisomal oxidative burnout and damage during endotoxin-induced stress. Autophagy. 2013;9(3):442–4.
Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–22.
Behl C. BAG3 and friends: co-chaperones in selective autophagy during aging and disease. Autophagy. 2011;7(7):795–8.
Gamerdinger M, et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901.
Carra S, et al. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283(3):1437–44.
Gamerdinger M, et al. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12(2):149–56.
Hishiya A, et al. BAG3 directly interacts with mutated alphaB-crystallin to suppress its aggregation and toxicity. PLoS One. 2011;6(3):e16828.
Volpicelli-Daley LA, et al. Addition of exogenous alpha-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous alpha-synuclein to Lewy body and Lewy neurite-like aggregates. Nat Protoc. 2014;9(9):2135–46.
Hagemann TL, et al. Suppression of GFAP toxicity by alphaB-crystallin in mouse models of Alexander disease. Hum Mol Genet. 2009;18(7):1190–9.
Gallardo G, et al. A molecular pathway of neurodegeneration linking alpha-synuclein to ApoE and Abeta peptides. Nat Neurosci. 2008;11(3):301–8.
Tannous P, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105(28):9745–50.
Carver JA, et al. Small heat-shock proteins and clusterin: intra- and extracellular molecular chaperones with a common mechanism of action and function? IUBMB Life. 2003;55(12):661–8.
Ecroyd H, Carver JA. Crystallin proteins and amyloid fibrils. Cell Mol Life Sci. 2009;66(1):62–81.
Masilamoni JG, et al. The protective effect of alpha-crystallin against acute inflammation in mice. Biochim Biophys Acta. 2005;1740(3):411–20.
Masilamoni JG, et al. The neuroprotective efficacy of alpha-crystallin against acute inflammation in mice. Brain Res Bull. 2005;67(3):235–41.
Ousman SS, et al. Protective and therapeutic role for alphaB-crystallin in autoimmune demyelination. Nature. 2007;448(7152):474–9.
Kuipers HF, et al. Phosphorylation of alphaB-crystallin supports reactive astrogliosis in demyelination. Proc Natl Acad Sci U S A. 2017;114(9):E1745–54.
Dewachter I, et al. Neuronal deficiency of presenilin 1 inhibits amyloid plaque formation and corrects hippocampal long-term potentiation but not a cognitive defect of amyloid precursor protein [V717I] transgenic mice. J Neurosci. 2002;22(9):3445–53.
Kallfass C, et al. Visualizing production of Beta interferon by astrocytes and microglia in brain of La Crosse virus-infected mice. J Virol. 2012;86(20):11223–30.
Consortium UKPsD, et al. Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20(2):345–53.
Vandrovcova J, et al. Association of MAPT haplotype-tagging SNPs with sporadic Parkinson’s disease. Neurobiol Aging. 2009;30(9):1477–82.
Wider C, et al. Association of the MAPT locus with Parkinson’s disease. Eur J Neurol. 2010;17(3):483–6.
Acknowledgments
We thank Ms. YD Li and JC Hou for excellent technical assistance in genotyping; Dr. Q Hu and his colleagues at the Optical Imaging Center of ION for technical support in confocal microscopy; Dr. Y Kong and her colleagues at the EM core-facility of ION for technical support in electron microscopy; Drs. VM Lee, J Trojanowski and KC Luk for providing α-synuclein PFFs.
Funding
This work was supported by grants from the Natural Science Foundation of China (31430036, 91742116, U1801681), National Key Basic Research Program of China (2015CB553500), Key Research Program of Frontier Sciences (QYZDJ-SSW-SMC002), Strategic Priority Research Program of Chinese Academy of Science (XDB32020100), Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), and the Shanghai Municipal Science and Technology Commission (17ZR1435300 to SZZ).
Availability of data and materials
The data supporting the conclusions of this article are available from the corresponding author upon request.
Author information
Authors and Affiliations
Contributions
SZL and YSG conducted most of the in vivo and in vitro experiments and the data analysis. PZL contributed to pilot experiments. SZZ and SY contributed to in vivo experiments. YQY contributed to cell cultures. XMW, FD and XSG contributed to discussion. JWZ supervised the project and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures performed were approved by the Institutional Animal Care and Use Committee and were in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lu, Sz., Guo, Ys., Liang, Pz. et al. Suppression of astrocytic autophagy by αB-crystallin contributes to α-synuclein inclusion formation. Transl Neurodegener 8, 3 (2019). https://doi.org/10.1186/s40035-018-0143-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40035-018-0143-7