Abstract
Introduction
While COVID-19 pandemic continues to spread worldwide, researchers have linked patterns of traits to poor disease outcomes. Risk factors for COVID-19 include asthma, elderly age, being pregnant, having any underlying diseases such as cardiovascular disease, diabetes, obesity, and experiencing lifelong systemic racism. Recently, connections to certain genes have also been found, although the susceptibility has not yet been established. We aimed to investigate the available evidence for the genetic susceptibility to COVID-19.
Methods
This study was a systematic review of current evidence to investigate the genetic susceptibility of COVID-19. By systematic search and utilizing the keywords in the online databases including Scopus, PubMed, Web of Science, and Science Direct, we retrieved all the related papers and reports published in English from December 2019 to September 2020.
Results
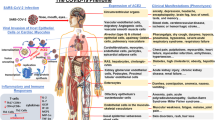
According to the findings, COVID-19 uses the angiotensin-converting enzyme 2 (ACE2) receptor for cell entry. Previous studies have shown that people with ACE2 polymorphism who have type 2 transmembrane serine proteases (TMPRSS2) are at high risk of SARS-CoV-2 infection. Also, two studies have shown that males are more likely to become infected with SARS-CoV-2 than females. Besides, research has also shown that patients possessing HLA-B*15:03 genotype may become immune to the infection.
Conclusion
Combing through the genome, several genes related to immune system’s response were related to the severity and susceptibility to the COVID-19. In conclusion, a correlation was found between the ACE2 levels and the susceptibility to SARS-CoV-2 infection.
Similar content being viewed by others
Introduction
Coronavirus Disease-2019 (COVID-19) started its devastating trajectory into a global pandemic in Wuhan, China [1,2,3,4,5,6]. The World Health Organization (WHO) declared this disease a “public health emergency of international concern” on January 30, 2020 [7,8,9]. Until April 1, the WHO reports a total number of 127,877,462 confirmed cases and 2,796,561 deaths [10]. Although capable of causing severe pneumonia, severe acute respiratory syndrome-Coronavirus-2 (SARS-CoV-2) can also involve various organs and cause physical symptoms, as well as psychological damages [11,12,13,14].
SARS-CoV-2 utilizes molecules to enter the cells, such as angiotensin-converting enzyme 2 (ACE2) to attach to the receptor-binding domain (RBD) and type 2 transmembrane serine proteases (TMPRSS2) to cleave the spike (S) protein [15,16,17]. SARS-CoV-2 S-protein cleavage by TMPRSS2 initiates viral entry and also helps the virus escape the immune system [15]. The hidden receptor-binding domain attached to ACE2 enhances immune evasion by weakening the immune surveillance. SARS-CoV-2 S-protein cleavage by TMPRSS2 is also responsible for immune escape besides initiating the viral entry [15]. Hence, genetic differences among such molecules responsible for cellular entry might alter the observed responses among different individuals [18, 19]. Various cells in the body express ACE2 to different extents; and therefore, numerous organs of the body can be invaded by the virus, such as the lungs and the heart [18, 20,21,22]. Additionally, cell entry is reinforced in the presence of TMPRSS2 in ACE2-positive cells by cleaving the viral Spike protein, which can result in its activation for membrane fusion [23,24,25]. There are reports that not only genetic differences can cause certain races and populations to be more affected by this virus, but also they might contribute to gender-specific differences [26]. Higher testosterone levels enhance TMPRSS2 levels and might cause higher disease susceptibility in males [27, 28]. To make matters even more important, the differences in severity observed in children compared to older adults may be in parts due to the different expressions of genetic components, such as TMPRSS2 [29].
As mentioned above, various genetic patterns might alter the risk of infection, viral entry, immune response to the virus, COVID-19 severity, and death. Therefore, we aim to review the published literature on the genetic aspects of susceptibility to COVID-19 and discuss the above-mentioned statements. Recognizing these points might help us determine at-risk groups and take specific measures to support them.
Methods
This study aims to review the current evidence of genetic susceptibility to COVID-19 carried out in September 2020. The authors studied the probable genetic vulnerability to novel Coronavirus (COVID-19). In this study, the preferred reporting items for systematic reviews and meta-analyses (PRISMA) checklist is used as a criterion to ensure the accuracy and reliability of the results.
Data sources
By systematic search and utilizing the keywords in the online databases including Scopus, PubMed, Web of Science, and Science Direct, we retrieved all related papers and reports published in English from December 2019 to September 2020. To convey our search strategy, we considered different combinations of keywords in the following orders:
-
1.
“COVID-19” or “SARS-CoV-2” or “Novel Coronavirus” or “2019-nCoV” or “Coronavirus” (title/abstract).
-
2.
“Genetic susceptibility” or “Genetic vulnerability” or “Genetic probability” (title/abstract).
-
3.
(A) and (B).
Study selection
Two independent investigators screened and selected the studies with the most relevant titles and abstracts. Then, the full text of the extracted papers was reviewed, and based on the eligibility criteria, the most relevant papers were selected. The peer-reviewed original papers published in English that satisfied the eligibility criteria were considered in the final report.
Our exclusion criteria for this study, whereas the following measures:
-
Papers focusing on non-human studies including in vitro observations or researches concentrating on animal experiments, or discussing COVID-19 without a satisfying detail, and papers without referring to the keywords of this study.
-
Papers whose full texts were not accessible.
-
Any results were suspicious of duplication in databases.
Data extraction
After the summarization process, data including type of article (e.g., case reports), information of authors, country of origin, publication date, sample size, gender, age, and genetic susceptibility data were transferred into an organized sheet. Two independent investigators gathered this information. Finally, the other authors cross-checked all the selected papers to make sure no overlap or duplications exist in the content.
Quality assessment
As previously mentioned, we considered the preferred reporting items for systematic reviews and meta-analyses (PRISMA) checklist to ensure the quality and authenticity of selected studies. The quality and consistency of the articles and the probable risk of bias were evaluated by two independent researchers. A third independent researcher was recruited to resolve the issue in any situation where differences in thoughts were encountered. In the end, the full text of selected articles was read and the significant findings of each article were extracted.
Results
Using the described search strategy, we retrieved 148 full-text articles. After an initial review of retrieved resources, 69 duplicates were removed, and the title and abstract of the remaining 79 articles were reviewed. Applying the selection criteria, 58 articles were excluded, and only 21 articles met the inclusion criteria and were included in the final review (Fig. 1).
According to the findings, COVID-19 uses the ACE2 receptor for cell entry. A review of articles showed individuals with ACE2 polymorphism along with TMPRSS2 are more vulnerable to COVID-19. Two studies have shown that males are more likely to become infected with SARS-CoV-2 than females. Besides, research have also shown that patients possessing HLA-B*15:03 genotype may become immune to the infection (Table 1).
Discussion
Consistent with the general conclusion, there are several genetic patterns of susceptibility for COVID-19, including: ACE2, TMPRSS2, Cathepsin, FURIN, MX dynamin-like GTPase 1 (MX1), Basigin (BSG), IFN-γ, IL-17A/F, HLA, cytokine storm, TLR genes, p.Ser1103, Tyr-SCN5A, p.Arg514Gly, p.Val160Met, p.Gly181Arg, p.Arg240Cys, p.Gly259Ser, p.Pro335Leu, p.Gly432Ala, Telomeres, and immune complexes (ICs) [18, 19, 30, 32, 33, 38,39,40, 43, 44, 51]. Our review demonstrated that genetic risk factors can affect the treatment and prevention of COVID-19.
ACE2 and TMPRSS2 DNA polymorphisms have been shown to be strongly associated with the susceptibility, severity, and clinical outcomes of COVID-19 [18, 19]. Structural analysis of the binding interface between SARS-CoV-2 RBD and human ACE2 shows a strong network of polar contacts [46]. SARS-CoV-2 RBD mediates these polar interactions through Lys417, Gly446, Tyr449, Asn487, Gln493, Gln498, Thr500, Asn501, Gly502 and Tyr505 with ACE2 [46]. Various gender and age groups have significantly different susceptibilities to infection and mortality rates [52]. Old age males with comorbidities are considered the most vulnerable groups [27, 28, 30, 52]. SARS-CoV-2 has a very strong interaction with the ACE2 receptor, and it has an important role in cell entry together with TMPRSS2 [18, 19, 52,53,54,55,56,57]. Males are at higher risk of severe COVID-19 compared to women and are responsible for 60% of the deaths [30, 52]. The ACE2 expression rate is higher in females than males, single nucleotide polymorphisms (SNPs) within the TMPRSS2 gene (21q22.3) can also have a greater role in the general population (rs2070788, rs7364083, rs9974589) and in a sex-oriented perspective (rs8134378) assuming that higher expression in males might favor virus membrane fusion [26, 37].
ACE2 is a receptor for SARS-CoV-2. Also, it can facilitate the post-infection downstream processes including inflammatory responses [33]. ACE2 receptors are ubiquitous and widely expressed in the heart, vessels, gut, lung, kidney, testis, and brain [20, 22, 58]. SARS-CoV-2 enters into the cell through binding of the viral S-protein to the ACE2 receptors [15, 17]. The ACE2 receptors diminish the unfavorable effects of angiotensin II in several ways [58]. First, they can degrade angiotensin II and reduce its deleterious effect, and second, by generating angiotensin (1–7) they perform counter-regulatory effects to those of angiotensin II [59]. Several conditions including older age, diabetes, hypertension, and heart diseases like heart failure can lead to ACE2 deficiency [58]. Given the above information, ACE2 deficiency is supposed to play a pivotal role in the pathogenesis of SARS-CoV-2 infection which favors the progression of inflammatory and thrombotic processes [58].
As mentioned earlier, binding SARS-CoV-2 to ACE2 is facilitated by a RBD located within the S-protein of the virus. There are 25 different ACE2 gene variants in the Leiden open variation database (LOVD) [37]. Two specific ACE2 alleles (i.e., rs73635825 and rs143936283) showed a low binding affinity of the entire SARS-CoV-2 S-protein, so viral attachment decreased and potential resistance to infection was reported [37]. According to recent studies, genetic polymorphisms in the ACE2 gene were detected in populations, which modulate the SARS-CoV-2 affinity spike protein and worsen pulmonary and systemic injury by accelerating lung damage in COVID-19 patients, which enhanced vasoconstriction, inflammation, oxidation, coagulation, capillary permeability, fibrosis, and apoptosis in the alveolar cells [37, 60]. Single-nucleotide polymorphism (SNPs) in ACE2 is associated with different diseases. Therefore, SNPs can impact gene expression, which might influence disease severity. Moreover, COVID-19 is referred to the state of cytokine storm which results in an excessive release of cytokines that lead to multiple organ damage. Such response cytokine storm was showed in ACE2-positive cells. Thus, differences in response to COVID-19 associated with genetic polymorphism within the genes responsible for the synthesis of cytokines and pro-inflammatory mediators along with ACE2 [61].
Male express higher ACE2 levels in their type II pneumocytes; and therefore, their lung samples may express threefold higher levels of ACE2 compared to the females [53,54,55,56,57]. In addition, the ACE2 expression in Asian populations is higher than African-American and Caucasian populations [31, 34]. As the evidence showed, the conversion of angiotensin II to angiotensin (1–7) by ACE2 was higher in males than females, indicating over expression of ACE2 in males [54]. As ACE2 is encoded by a gene located on the X chromosome and males express more ACE2 than females it could be considered that depending on the allele expressed by females, they could be considered of lower sensitivity against the most severe adverse effects of the infection [54].
As thoroughly discussed above, ACE2 protein at the surface of lung alveolar epithelial cells allows infection of the respiratory tract with SARS-CoV-2 [54]. In the lung, SARS-CoV-2 mainly infects alveolar epithelial cells, resulting in respiratory symptoms, and SARS-CoV infects mainly pneumocytes and macrophages. ACE2 expression protects from lung injury and is down-regulated by SARS-S. The expression of ACE2 also manifests itself beyond the lung. Since ACE2 is a functional receptor for SARS-CoV-2, researchers started to carefully consider the safety and potential effects of antihypertension therapy with ACE inhibitors or angiotensin-receptor blockers in patients with COVID-19. Besides, diabetes mellitus and hypertension are known as the most common comorbidities in COVID-19; both these conditions are controlled by ACE2. Earlier researchers were concerned on whether patients with COVID-19 and hypertension who are taking an ACE inhibitor (ACEI) or angiotensin-receptor blocker (ARB) medication should seek another antihypertensive drug [11, 41]. However, novel studies refuted the association between ACEI/ARB use and the COVID-19 disease severity; and therefore, physicians should not have concerns regarding prescribing these medications during the pandemic [62, 63].
On the other hand, a TMPRSS2 inhibitor can be a treatment option by blocking the cell entry, since an inhibitor of TMPRSS2 can block SARS-CoV-2 infection of lung cells [18, 41].
ACE2 can cause SARS-CoV-2 infection in the host cells, while also causing damage to the myocardium; however, the specific mechanisms have not yet been precisely discovered. Some studies indicated that ACE2 polymorphisms were in association with cardiovascular conditions by modifying the angiotensinogen–ACE2 interactions, asp.Arg514Gly in the African/African-American people. Particularly, 54% (33/61) and 39% (24/61) of deleterious variants in ACE2 appear in non-Finnish European (EUR) and African/African-American (AFR) populations, respectively. One of the reasons for the difference in genetic susceptibility to COVID-19 on one hand, and risk factors like high-risk male patients with cancer, on the other hand, can be the presence of a high number of polymorphisms in TMPRSS2 [e.g., p.Val160Met (rs12329760) which is an expression quantitative trait locus (eQTL)]. Moreover, the comparative genetic analyses implied that ACE2 genomic variants play critical roles in susceptibilities to COVID-19 and its associated cardiovascular conditions by altering the AGT-ACE2 pathway. Also, the developmental regulation of TMPRSS2 may make infants and children immune from COVID-19. The prevalent polymorphisms in ACE2 or TMPRSS2 lead to the use of personalized treatments (i.e., hydroxychloroquine and camostat) for COVID-19. However, the evidence refuted the effectiveness of these treatments and ruled out their use as logical treatments [64]. Also TMPRSS2 variants and resulting expression influence COVID-19 severity [26, 65]. For example, endometrium carries low susceptibility to COVID-19 due to the low expression of TMPRSS2 and ACE2. However, expression of the genes related to SARS-CoV-2 infection varies with menstrual cycle phase and age, with older women expressing higher ACE2 levels [30].
Last but not least, the similarity of the chemokine receptor protein encoded by the CCR5 gene for HIV has been compared to ACE2 for SARS-CoV. The target of HIV infection is CCR5 gene, and genetic variations in CCR5 can result in the different rates of this infection. ACE2 can play the role of an efficient or inefficient receptor for SARS-CoV if the amino acid remains are altered. Several ACE2 variants could have an important influence on the association between ACE2 and S-protein in SARS-CoV. Therefore, this gene variation can justify the gender differences and different susceptibility and disease severity to SARS-CoV infection in different racial groups [36].
In addition, some evidence indicates that the entry of SARS-CoV-2 into the cell is facilitated by the polymorphisms of the cellular proteases in the presence of furin [65]. The furin can be connected to the SARS-CoV-2, and the S-protein might be cleaved by TMPSS2 with the collaboration of furin. This function results in the entry of the virus into the respiratory tract and also in an increased risk of infection. Despite the aforementioned procedures, an association between furin and COVID-19 has not yet been discovered [49].
Conclusion
Combing through the genome, a relation has been found between the severity and susceptibility of COVID-19 and some genes associated with the immune system’s response. In conclusion, a correlation was found between the ACE2 levels and the susceptibility to SARS-CoV-2 infection. Therefore, in SARS-CoV-2, as well as SARS-CoV, ACE2 has been recognized as a functional receptor. ACE2 seems to be crucial in outcomes of COVID-19, and the efficiency of ACE2 usage is a significant element of SARS-CoV transmissibility. SARS-2-S uses ACE2 (SARS-CoV receptor) to enter the cells and the serine protease TMPRSS2 for S-protein preparing to the entrance.
Availability of data and materials
The authors stated that all information provided in this article could be shared.
Abbreviations
- ACE2:
-
Angiotensin-converting enzyme 2
- AFR:
-
African/African-American
- BSG:
-
Basigin
- COVID-19:
-
Coronavirus disease-2019
- HIV:
-
Human immunodeficiency virus
- EUR:
-
Non-Finnish European
- PRISMA:
-
Preferred reporting items for systematic reviews and meta-analyses
- TMPRSS2:
-
Type 2 transmembrane serine proteases
- WHO:
-
World Health Organization
References
WHO. WHO director-general’s opening remarks at the media briefing on COVID-19. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19. Accessed on 11 Mar 2020.
SeyedAlinaghi S, Abbasian L, Solduzian M, Ayoobi Yazdi N, Jafari F, Adibimehr A, Farahani A, Salami Khaneshan A, Ebrahimi Alavijeh P, Jahani Z, Karimian E, Ahmadinejad Z, Khalili H, Seifi A, Ghiasvand F, Ghaderkhani S, Rasoolinejad M. Predictors of the prolonged recovery period in COVID-19 patients: a cross-sectional study. Eur J Med Res. 2021;26(1):41. https://doi.org/10.1186/s40001-021-00513-x.
Mehraeen E, Hayati B, Saeidi S, Heydari M, SeyedAlinaghi S. Self-care instructions for people not requiring hospitalization for Coronavirus disease 2019 (COVID-19). Arch Clin Infect Dis. 2020;15(COVID-19):e102978.
Sekhavati E, Jafari F, SeyedAlinaghi S, Jamalimoghadamsiahkali S, Sadr S, Tabarestani M, et al. Safety and effectiveness of azithromycin in patients with COVID-19: an open-label randomised trial. Int J Antimicrob Agents. 2020;56(4):106143.
SeyedAlinaghi S, Ghadimi M, Hajiabdolbaghi M, Rasoolinejad M, Abbasian L, Nezhad MH, et al. Prevalence of COVID-19-like symptoms among people living with HIV, and using antiretroviral therapy for prevention and treatment. Curr HIV Res. 2020;18:373.
Seyed ASA, Karimi A, Shobeiri P, Nowroozi A, Mehraeen E, Afsahi AM, et al. Psychological symptoms of COVID-19 epidemic: a systematic review of current evidence. Psihologija. 2020;54:35.
Ghiasvand F, Miandoab SZ, Harandi H, Golestan FS, Alinaghi SAS. A patient with COVID-19 disease in a referral hospital in Iran: a typical case. Infect Disord Drug Targets. 2020;20(4):559–62. https://doi.org/10.2174/1871526520666200429115535.
Mehraeen E, Behnezhad F, Salehi MA, Noori T, Harandi H, SeyedAlinaghi S. Olfactory and gustatory dysfunctions due to the Coronavirus disease (COVID-19): a review of current evidence. Eur Arch Otorhinolaryngol. 2020;278:1–6.
Ghiasvand F, Ghadimi M, Ghadimi F, Safarpour S, Hosseinzadeh R, SeyedAlinaghi S. Symmetrical polyneuropathy in Coronavirus disease 2019 (COVID-19). IDCases. 2020;21:e00815.
WHO. WHO Coronavirus disease (COVID-19) dashboard: https://covid19.who.int/. Accessed 01 Apr 2020.
Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020;17(5):259–60.
Sadr S, SeyedAlinaghi S, Ghiasvand F, Hassan Nezhad M, Javadian N, Hossienzade R, et al. Isolated severe thrombocytopenia in a patient with COVID-19: a case report. IDCases. 2020;21:e00820.
Ghiasvand F, SeyedAlinaghi S. Isolated anosmia as a presentation of COVID-19: an experience in a referral hospital. Infect Disord Drug Targets. 2020;20(3):350.
Ahmadinejad Z, Salahshour F, Dadras O, Rezaei H, Alinaghi S. Pleural effusion as a sign of Coronavirus disease 2019 (COVID-19) pneumonia: a case report. Infect Disord Drug Targets. 2020. https://doi.org/10.2174/1871526520666200609125045.
Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727.
Mollica V, Rizzo A, Massari F. The pivotal role of TMPRSS2 in Coronavirus disease 2019 and prostate cancer. Future Oncol. 2020;16(27):2029–33.
Deng Q, Rasool RU, Russell RM, Natesan R, Asangani IA. Targeting androgen regulation of TMPRSS2 and ACE2 as a therapeutic strategy to combat COVID-19. iScience. 2021;24(3):102254.
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-80e8.
Shirato K, Kawase M, Matsuyama S. Wild-type human Coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15.
Mehraeen E, Karimi A, Barzegary A, Vahedi F, Afsahi AM, Dadras O, et al. Predictors of mortality in patients with COVID-19—a systematic review. Eur J Integr Med. 2020;40:101226.
Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS Coronavirus. Nature. 2003;426(6965):450–4.
Mehraeen E, Seyed Alinaghi SA, Nowroozi A, Dadras O, Alilou S, Shobeiri P, et al. A systematic review of ECG findings in patients with COVID-19. Indian Heart J. 2020;72(6):500–7.
Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome Coronavirus spike protein. J Virol. 2014;88(2):1293–307.
Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome Coronavirus entry. J Virol. 2012;86(12):6537–45.
Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, et al. Influenza and SARS-Coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE. 2012;7(4):e35876.
Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):216.
Cattrini C, Bersanelli M, Latocca MM, Conte B, Vallome G, Boccardo F. Sex hormones and hormone therapy during COVID-19 pandemic: implications for patients with cancer. Cancers. 2020;12(8):2325.
Bennani NN, Bennani-Baiti IM. Androgen deprivation therapy may constitute a more effective COVID-19 prophylactic than therapeutic strategy. Ann Oncol. 2020;31(11):1585–6.
Schuler BA, Habermann AC, Plosa EJ, Taylor CJ, Jetter C, Kapp ME, et al. Age-related expression of SARS-CoV-2 priming protease TMPRSS2 in the developing lung. bioRxiv. 2020. https://doi.org/10.1101/2020.05.22.111187.
Abhari S, Kawwass JF. Endometrial susceptibility to SARS CoV-2: explained by gene expression across the menstrual cycle? Fertil Steril. 2020;114(2):255–6.
Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, et al. Comparative genetic analysis of the novel Coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6(1):1–4.
Casanova J-L, Su HC, Effort CHG. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194.
Debnath M, Banerjee M, Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020;34:8787.
Devaux CA, Rolain J-M, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53:425.
Fakhouri EW, Peterson SJ, Kothari J, Alex R, Shapiro JI, Abraham NG. Genetic polymorphisms complicate COVID-19 therapy: pivotal role of HO-1 in cytokine storm. Antioxidants. 2020;9(7):636.
Fujikura K, Uesaka K. Genetic variations in the human severe acute respiratory syndrome Coronavirus receptor ACE2 and serine protease TMPRSS2. J Clin Pathol. 2020;74:307.
Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int J Mol Sci. 2020;21(10):3474.
Giudicessi JR, Roden DM, Wilde AA, Ackerman MJ. Genetic susceptibility for COVID-19-associated sudden cardiac death in African Americans. Heart Rhythm. 2020;17:1487.
Godri Pollitt KJ, Peccia J, Ko AI, Kaminski N, Dela Cruz CS, Nebert DW, et al. COVID-19 vulnerability: the potential impact of genetic susceptibility and airborne transmission. Hum Genomics. 2020;14:1–7.
Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):1–8.
Junejo Y, Ozaslan M, Safdar M, Khailany RA, Rehman S, Yousaf W, et al. Novel SARS-CoV-2/COVID-19: origin, pathogenesis, genes and genetic variations, immune responses and phylogenetic analysis. Gene Rep. 2020;20:100752.
Kachuri L, Francis SS, Morrison M, Bossé Y, Cavazos TB, Rashkin SR, et al. The landscape of host genetic factors involved in infection to common viruses and SARS-CoV-2. medRxiv. 2020;10:59.
LoPresti M, Beck DB, Duggal P, Cummings DA, Solomon BD. The role of host genetic factors in Coronavirus susceptibility: review of animal and systematic review of human literature. medRxiv. 2020;3:35.
Sanchez-Gonzalez MA, Moskowitz D, Issuree PD, Yatzkan G, Rizvi S, Day K. A pathophysiological perspective on COVID-19’s lethal complication: from viremia to hypersensitivity pneumonitis-like immune dysregulation. Infect Chemother. 2020;52:e31.
Sheikh JA, Singh J, Singh H, Jamal S, Khubaib M, Kohli S, et al. Emerging genetic diversity among clinical isolates of SARS-CoV-2: lessons for today. Infect Genet Evol. 2020;84:104330.
Sironi M, Hasnain SE, Phan T, Luciani F, Shaw M-A, Sallum MA, et al. SARS-CoV-2 and COVID-19: a genetic, epidemiological, and evolutionary perspective. Infect Genet Evol. 2020;84:104384.
Lopera Maya EA, van der Graaf A, Lanting P, van der Geest M, Fu J, Swertz M, et al. Lack of association between genetic variants at ACE2 and TMPRSS2 genes involved in SARS-CoV-2 infection and human quantitative phenotypes. Front Genet. 2020;11:613.
Strafella C, Caputo V, Termine A, Barati S, Gambardella S, Borgiani P, et al. Analysis of ACE2 genetic variability among populations highlights a possible link with COVID-19-related neurological complications. Genes. 2020;11:741.
Torre-Fuentes L, Matías-Guiu J, Hernández-Lorenzo L, Montero-Escribano P, Pytel V, Porta-Etessam J, et al. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J Med Virol. 2020;93:863.
von der Thüsen J, van der Eerden M. Histopathology and genetic susceptibility in COVID-19 pneumonia. Eur J Clin Investig. 2020. https://doi.org/10.1111/eci.13259.
Vuitton DA, Vuitton L, Seillès E, Galanaud P. A plea for the pathogenic role of immune complexes in severe COVID-19. Clin Immunol. 2020;217:108493.
Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Norman L, et al. Features of 20 133 UK patients in hospital with COVID-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ. 2020;369:m1985.
Baker SA, Kwok S, Berry GJ, Montine TJ. Angiotensin-converting enzyme 2 (ACE2) expression increases with age in patients requiring mechanical ventilation. PLoS ONE. 2021;16(2):e0247060.
Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–35.
Song H, Seddighzadeh B, Cooperberg MR, Huang FW. Expression of ACE2, the SARS-CoV-2 receptor, and TMPRSS2 in prostate epithelial cells. bioRxiv. 2020. https://doi.org/10.1101/2020.04.24.056259.
Beyerstedt S, Casaro EB, Rangel ÉB. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021;40:905.
Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202(5):756–9.
Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14.
Santos RA, Ferreira AJ, Verano-Braga T, Bader M. Angiotensin-converting enzyme 2, angiotensin-(1–7) and Mas: new players of the renin–angiotensin system. J Endocrinol. 2013;216(2):R1-17.
Lippi G, Lavie CJ, Henry BM, Sanchis-Gomar F. Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in Coronavirus disease 2019 (COVID-19)? Clin Chem Lab Med. 2020;58:1415.
Chaudhary M. COVID-19 susceptibility: potential of ACE2 polymorphisms. Egypt J Med Human Genet. 2020;21(1):1–8.
Fosbøl EL, Butt JH, Østergaard L, Andersson C, Selmer C, Kragholm K, et al. Association of angiotensin-converting enzyme inhibitor or angiotensin receptor blocker use with COVID-19 diagnosis and mortality. JAMA. 2020;324(2):168–77.
Kuster GM, Pfister O, Burkard T, Zhou Q, Twerenbold R, Haaf P, et al. SARS-CoV2: should inhibitors of the renin–angiotensin system be withdrawn in patients with COVID-19? Eur Heart J. 2020;41(19):1801–3.
Furtado RHM, Berwanger O, Fonseca HA, Corrêa TD, Ferraz LR, Lapa MG, et al. Azithromycin in addition to standard of care versus standard of care alone in the treatment of patients admitted to the hospital with severe COVID-19 in Brazil (COALITION II): a randomised clinical trial. Lancet. 2020;396(10256):959–67.
Godri Pollitt KJ, Peccia J, Ko AI, Kaminski N, Dela Cruz CS, Nebert DW, et al. COVID-19 vulnerability: the potential impact of genetic susceptibility and airborne transmission. Hum Genom. 2020;14(1):17.
Acknowledgements
The present study was conducted in collaboration with Khalkhal University of Medical Sciences, Iranian Institute for Reduction of High-Risk Behaviors, Tehran University of Medical Sciences, and Department of Global Health and Socioepidemiology, Kyoto University.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
The conception and design of the study: EM, SS. Acquisition of data: AK, BM-B, PH. Analysis and interpretation of data: PM, MM, AB. Drafting the article: EM, MM, AMA, MH. Revising it critically for important intellectual content: SS, SJ. Final approval of the version to be submitted: EM, OD, J-MS, FV. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was extracted from the research project with code IR.KHALUMS.REC.1399.001 entitled “Investigation of effective drugs for people affected by Coronavirus disease 2019 (COVID-19) in selected hospitals in Ardabil” conducted at Khalkhal University of Medical Sciences in 2020.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests regarding the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
SeyedAlinaghi, S., Mehrtak, M., MohsseniPour, M. et al. Genetic susceptibility of COVID-19: a systematic review of current evidence. Eur J Med Res 26, 46 (2021). https://doi.org/10.1186/s40001-021-00516-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-021-00516-8