Abstract
Resveratrol is a typical plant phenolic compound whose derivatives are synthesized through hydroxylation, O-methylation, prenylation, and oligomerization. Resveratrol and its derivatives exhibit anti-neurodegenerative, anti-rheumatoid, and anti-inflammatory effects. Owing to the diverse biological activities of these compounds and their importance in human health, this study attempted to synthesize five resveratrol derivatives (isorhapontigenin, pterostilbene, 4-methoxyresveratrol, piceatannol, and rhapontigenin) using Escherichia coli. Two-culture system was used to improve the final yield of resveratrol derivatives. Resveratrol was synthesized in the first E. coli cell that harbored genes for resveratrol biosynthesis including TAL (tyrosine ammonia lyase), 4CL (4-coumaroyl CoA ligase), STS (stilbene synthase) and genes for tyrosine biosynthesis such as aroG (deoxyphosphoheptonate aldolase) and tyrA (prephenate dehydrogenase). Thereafter, culture filtrate from the first cell was used for the modification reaction carried out using the second E. coli harboring hydroxylase and/or O-methyltransferase. Approximately, 89.8 mg/L of resveratrol was synthesized and using the same, five derivatives were prepared with a conversion rate of 88.2% to 22.9%. Using these synthesized resveratrol derivatives, we evaluated their anti-inflammatory activity. 4-Methoxyresveratrol, pterostilbene and isorhapontigenin showed the anti-inflammatory effects without any toxicity. In addition, pterostilbene exhibited the enhanced anti-inflammatory effects for macrophages compared to resveratrol.
Similar content being viewed by others
Introduction
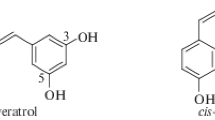
Resveratrol is a natural phenolic compound that belongs to the stilbenoid group. Resveratrol and its glucoside (resveratrol 3-O-glucoside, piceid) are the two major stilbenoids found in nature [1]. Grape is one of the primary sources of resveratrol and its derivatives [2]. Major resveratrol derivatives are synthesized via hydroxylation, glycosylation, O-methylation, and prenylation [3]; resveratrol derivatives from oligomerization have been reported in plants [4]. Resveratrol is synthesized from tyrosine through deamination by tyrosine ammonia lyase (TAL), Co-A attachment by 4-hydroxycinnamoyl-CoA ligase (4CL), and condensation with three molecules of malonyl-CoA by stilbene synthase (STS) [5]. Genes that encode each step of resveratrol biosynthesis from tyrosine have been cloned and characterized [3]. Among them, gene encoding STS, a type III polyketide synthase [6], has been cloned in various plants, including Arachis hypogaea (groundnut; [7]), Pinus densiflora [8], and Vitis vinifera [6]. Although plants are known to be a good source, resveratrol has been synthesized in a microbial system by introducing genes associated with biosynthesis (reviewed by [3]). Previous studies included selection of the best combination of TAL, STS, and 4CL, and supplementation with additional p-coumaric acid and malonyl-CoA [3]. These approaches included pathway engineering to enhance the supplementation of tyrosine and malonyl-CoA or modulation of the entire pathway. Using Saccharomyces cerevisiae, 415.7 mg/L of resveratrol was synthesized from glucose via tyrosine and malonyl-CoA overproduction in fed-batch fermentation [9]. Implementing the pull–push block strategy, final yield of resveratrol in S. cerevisiae increased up to 800 mg/L [10]. In Escherichia coli, the resveratrol synthesis pathway was introduced, following different strategies, to increase the pool of malonyl-CoA; the resulting strains produced 304.5 mg/L resveratrol from glucose [11] and 238.71 mg/L resveratrol from tyrosine [12]. Not only resveratrol but also its derivatives could be synthesized, given the regioselective modification enzymes are available. Based on the structure of resveratrol, biological modifications are possible through O-methylation and hydroxylation. The three possible modification products include mono-O-methylation, di-O-methylation, or three-O-methylation. To date, E. coli has been engineered to produce 34.0 mg/L of pinostilbene (3-methylated resveratrol) and 0.16 mg/L of pterostilbene (3′,5′-dimethylated resveratrol) from 1 mM resveratrol [13]. Mixture of mono-, di-, and tri-O-methylated resveratrol was synthesized from glucose in E. coli [14]. On supplementing the culture medium with methionine to enhance the supply of S-adenosyl-L-methionine, 33.6 mg/L of pterostilbene was synthesized in E. coli [15]. Biosynthesis of pinostilbene (5.5 mg/L) and pterostilbene (34.9 mg/L) was also achieved in S. cerevisiae [10]. Piceatannol was synthesized from glucose in E. coli with a yield of 21.5 mg/L [16]. Overexpression of acetate and malonate assimilation pathways along with additional supply of malonate in the culture medium yielded 124 mg/L of piceatannol (3-hydroxyresveratrol) in E. coli [17].
Extensive research on the derivatives of resveratrol stems from their biological significance. Studies demonstrated that resveratrol downregulated the inflammatory response [18, 19] leading to decreased inflammation [20, 21]. This anti-inflammatory effect of resveratrol was considered one of the potential therapeutic strategies for treating various chronic diseases, such as cancer, diabetes, and neurodegenerative and cardiovascular disorders (reviewed by [19]).
Resveratrol derivatives that were modified by O-methylation and/or hydroxylation showed enhanced biological effects compared to resveratrol. Pterostilbene was reported to exhibit anticancer, anti-inflammatory, and antioxidant effects [22,23,24]. It also increasingly inhibited induction of inflammatory genes, such as inducible nitric oxide synthase and cyclooxygenase 2 [23]. Apart from the antioxidative and antiproliferative activities [25], piceatannol also demonstrated anticancer effect [26]. In addition, this compound was studied for its anti-inflammatory effects as well [27,28,29]. Rhapontigenin demonstrated anticancer, anti-inflammatory, cardioprotective, antiallergic, and antithrombotic effects [24, 30,31,32], while isorhapontigenin (3-methoxyresveratrol) showed anticancer [33], antioxidative [34], and anti-inflammatory activities. Compared to resveratrol, oral bioavailability of isorhapontigenin was much higher [35].
Resveratrol derivatives are highly valued owing to their diverse biological activities. Hence, in this study, attempt was made to synthesize resveratrol and five resveratrol derivatives using E. coli (Fig. 1). Simple modification reactions found in nature were mimicked by introducing the corresponding genes into a heterologous system. In this regard, the two-culture system was found to be effective for the modification reactions; one for resveratrol biosynthesis and the other for modification of resveratrol to obtain resveratrol derivatives. In addition, anti-inflammatory activities of the synthesized resveratrol derivatives were also evaluated.
Materials and methods
Materials
Lipopolysaccharides (LPS) from Pseudomonas aeruginosa 10 was purchased from Sigma Aldrich (St. Louis, MO, USA). The mouse tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) enzyme-linked immunosorbent assay (ELISA) kits were purchased from BD Biosciences (Franklin Lake, NJ, USA). Dulbecco’s modified Eagle’s medium (DMEM), Dulbecco’s phosphate-buffered saline (DPBS), penicillin/streptomycin (P/S), and fetal bovine serum (FBS) were purchased from Gibco BRL (Grand Island, NY, USA). A micro-bicinchoninic acid (BCA) protein assay kit was purchased from Thermo Fisher Scientific (Rockford, IL, USA).
Constructs
STS (DQ406692) was cloned from Vitis vinifera using reverse transcription polymerase chain reaction (RT-PCR). The primers used were ATGAATTCGATGGCTTCAGTCGAGGAATTT (EcoRI site is underlined) and ATGTCGACTTAATTTGTAACCATAGGAATG (SalI site is underlined). The PCR product was sequenced and subcloned into the EcoRI/SalI sites of pCDF-Duet (pC-VvSTS). R4CL [36] containing the ribosomal binding site and SalI site at the 5′ end and NotI site was subcloned into SalI/NotI site of pC-VvSTS. The resulting construct was named pC-VvSTS-Os4CL. SeTAL was cloned previously in NdeI/KpnI site of pCDF-Duet1 [37]. Using pC-VvSTS-Os4CL as a template, PCR was carried out with forward primer of VvSTS and reverse primer of Os4CL containing HindIII. The resulting PCR product was digested with EcoRI/HindIII and subcloned into EcoRI/HindIII of pC-SeTAL (pC-VvSTS-Os4CL-SeTAL).
ROMT15 and HpaBC were cloned previously [38, 39]. ROMT15 was subcloned into EcoRI/NotI sites of ROMT15 (pE-ROMT15) and HpaBC was subcloned into NdeI/XhoI site of pE-ROMT15 (pE-ROMT15-HpaBC). pG-ROMT9 and pG-SOMT2 were made previously [40, 41]. pE-SOMT2-HpaBC was constructed in a way similar to that of pE-ROMT15-HpaBC. Other genes used in this study were cloned previously [42,43,44] and reconstructed for use in the current study. Briefly, an internal BamHI site of tyrAf was eliminated by site-directed mutagenesis, without changing the amino acid sequence. It was then subcloned into NdeI/KpnI site of pACYC-duet-1 (pA-tyrAf). Other genes (aroG, aroGf, aroGf-ppsA-tktA, aroL-aroGf-ppsA-tktA, aroC-aroA-aroL-aroE-aroD-aroB-aroGf-ppsA-tktA, or aroC-aroA-aroL-ydiB-aroD-aroB-aroGf-ppsA-tktA) were subcloned into the BamHI/NotI sites of pA-tyrAf. The constructs and E. coli strains used in this study were listed in Table 1.
Synthesis of resveratrol derivatives
E. coli BL21 (DE3) was used as the host for synthesis of resveratrol derivatives. E. coli transformant was grown in Luria–Bertani (LB) broth with proper antibiotic(s) at 37 °C for 18 h. The culture was then inoculated into a fresh medium and allowed to grow at 37 °C until an optical density value of 1 at 600 nm was achieved. Cells were again resuspended in M9 medium containing 1% yeast extract and incubated at 30 °C for 24 h. The culture was extracted with three volumes of ethyl acetate. After being harvested, dried, and dissolved in dimethyl sulfoxide, the organic layer was analyzed using high performance liquid chromatography (HPLC). For the modification reaction of resveratrol, culture from the B-RS2 strain, initially used for resveratrol production (See Results and Discussion), was centrifuged. The supernatant was then harvested and boiled to destroy the antibiotics. Thereafter, it was mixed with the E. coli transformant harboring the construct for synthesis of resveratrol derivative. The reaction product was analyzed using HPLC, as described earlier [45]. All experiments were repeated thrice. Resveratrol was used to quantify the synthesized derivatives.
In order to determine the structures of the synthesized resveratrol derivatives, they were purified using thin layer chromatography (TLC; TLC silica gel 60 F254; Millipore, Burlington, MA, USA). The developing solvents used for each compound were as follows: isorhapontigenin (benzene: methanol: acetic acid = 6: 1.5: 0.1), pterostilbene (ethyl acetate: hexane: acetic acid = 1: 1: 0.1), 4-methoxyresveratrol (benzene: methanol = 5: 1), piceatannol (benzene: methanol: acetic acid = 15: 2.5: 0.1), and rhapontigenin (benzene: methanol: acetic acid = 8.5: 1: 0.1). Proton nuclear magnetic resonance spectroscopy (NMR) was carried out as described previously [46]. 1H NMR of isorhapontigenin in methanol-d6: δ 3.90 (3H, s), 6.16 (1H, dd, J = 2.1, 2.1 Hz), 6.46 (2H, d, J = 2.1 Hz), 6.77 (1H, d, J = 8.2 Hz), 6.83 (1H, d, J = 16.2 Hz), 6.95 (1H, dd, J = 8.2, 1.8 Hz), 6.96 (1H, d, J = 16.2 Hz), and 7.11 (1H, d, J = 1.8 Hz); 1H NMR of pterostilbene in chloroform-d1: δ 3.83 (6H, 3′/5′-methoxy, s), 6.38 (1H, H-4′, dd, J = 2.2, 2.2 Hz), 6.65 (2H, H-2′/H-6′, d, J = 2.2 Hz), 6.63 (2H, H-3/H-5, d, J = 9.3 Hz), 6.89 (1H, H-β, d, J = 16.3 Hz), 7.02 (1H, H-α, d, J = 16.3 Hz), and 7.39 (2H, H-2/H-6, d, J = 9.3 Hz); 1H NMR of 4-methoxyresveratrol in methanol-d4 (in ppm): δ 3.80 (3H, s), 6.16 (1H, dd, J = 2.1, 2.1 Hz), 6.45 (2H, d, J = 2.1 Hz), 6.84 (1H, d, J = 16.3 Hz), 6.89 (2H, d, J = 8.7 Hz), 6.98 (1H, d, J = 16.3 Hz), and 7.44 (2H, d, J = 8.9 Hz); 1H NMR of piceatannol in deuterated methanol: δ (ppm) 6.97 (d, J = 1.9 Hz, 1H), 6.88 (d, J = 16.2 Hz, 1H), 6.83 (dd, J = 2.0, 8.2 Hz, 1H), 6.74 (d, J = 15.2 Hz, 1H), 6.73 (d, J = 8.2 Hz, 1H), 6.42 (d, J = 2.1 Hz, 2H), and 6.15 (d, J = 2.1 Hz, 1H); 1H NMR of rhapontigenin in methanol-d4 (in ppm): δ 3.86 (3H, s, 4-OCH3), 6.17 (1H, dd, J = 2.1, 2.1 Hz, H4′), 6.45 (2H, d, J = 2.1 Hz, H2′/6′), 6.79 (1H, d, J = 16.1 Hz, Hβ), 6.89 (1H, d, J = 8.4 Hz, H5), 6.91 (1H, d, J = 16.1 Hz, Hα), 6.94 (1H, dd, J = 8.4, 2.0 Hz, H6), and 7.01 (1H, d, J = 2.0 Hz, H2).
Anti-inflammatory activity
RAW264.7 cells (murine macrophage cell line) were maintained in complete DMEM supplemented with 10% FBS, 100 U/mL of penicillin, and 100 μg/mL of streptomycin at 37 °C in a humidified atmosphere with 5% carbon dioxide. The cells were plated in 6-well plates at a density of 6 × 105 cells per well and incubated for 24 h prior to treatment. According to a previous study, the cells were pre-treated for 6 h with resveratrol and its derivatives at a concentration of 10 μM. Cells cultured in a complete medium were then treated with LPS (1 μg/mL) for 18 h in the presence of resveratrol derivatives at the same concentration [47]. Cells treated with LPS (1 μg/mL) alone were used as control. Cell viability was determined based on the amount of protein in the cells [48]. Briefly, after incubation, the cells were washed with DPBS and lysed using a lysis buffer. The cell lysate was centrifuged at 13,000 rpm for 5 min. The total amount of protein in the supernatant was quantified using a BCA protein assay kit, according to the manufacturer’s protocol. In order to determine the released cytokine level, supernatants were centrifuged at 13,000 rpm for 10 min at 4 °C to remove the remaining cell debris. Amounts of TNF-α and IL-6 in the supernatants were quantified using an ELISA assay kit, according to the manufacturer’s protocol. The data and error bars represent the mean values of independent measurements and the standard deviations for each experiment. Statistical analysis was performed using Student’s t-test. A p-value < 0.05 was considered statistically significant (95% confidence level).
Results and discussion
Optimization of the construct for resveratrol biosynthesis
Three genes, namely TAL, 4CL, and STS, synthesize resveratrol from tyrosine. E. coli was transformed with a construct containing all the three genes and the resulting transformant was tested for resveratrol synthesis. The synthesized compound showed a retention time similar to that of resveratrol, indicating successful synthesis of resveratrol. Approximately, 32.0 mg/L of resveratrol was synthesized after 24 h of reaction time. Absence of p-coumaric acid indicated that there could be a possibility for increased resveratrol synthesis, provided additional p-coumaric acid was added to the medium. Recent report also showed that these three genes were critical for the synthesis of resveratrol [49].
Since p-coumaric acid is synthesized from tyrosine, the amount of endogenous tyrosine plays a critical role in determining the final yield of resveratrol. Previously, it has been shown that overexpression of the genes in the shikimate pathway increases the yield by utilizing the intermediates in the shikimate pathway [43, 44, 50]. Herein, seven constructs with different combination of genes in shikimate pathway were tested. Each of the constructs was transformed along with pC-VvSTS-Os4CL-SeTAL into E. coli BL21 (DE3) cell. The resulting transformant (B-RS1 ~ 7) was then monitored for resveratrol synthesis. Results revealed that the culture filtrate of each strain not only contains resveratrol but also has p-coumaric acid. All the E. coli strains, except for the one harboring only pC-VvSTS-Os4CL-SeTAL, exhibited enhanced resveratrol synthesis. Among them, the B-RS3 strain reported highest resveratrol (89.8 mg/L) production and contained two feedback inhibition free genes (aroGf and tyrAf). The B-RS5 strain synthesized approximately 81.9 mg/L of resveratrol (Fig. 2). The final yield in B-RS3 was higher than that of the study using mutant strain [49].
Comparison of different module constructs for the synthesis of resveratrol. Each strain had genes encoding tyrosine ammonia lyase, 4-hydroxycinnamoyl-CoA ligase, and stilbene synthase. The different shikimate pathway gene modules indicated in Table 1 were introduced
In order to synthesize 4-hydroxycoumarin, quinoline, acridone, and 4-hyroxybenzoic acid in E. coli, we used module constructs that were prepared using various combination of genes involved in the shikimate pathway. Balance between the substrate(s) and product resulted in the highest yield. The level of p-coumaric acid, a substrate of resveratrol, was highest in the B-RS4 strain, followed by B-RS5 and B-RS3 strains. These three strains showed the highest resveratrol production. However, the best module construct for resveratrol production was different from that for other compound synthesis. This is likely to stem from the differences in substrate conversion capabilities of the downstream gene. The final yield of the current study was not comparable to the previous studies [11, 12]. Earlier E. coli was engineered to increase not only tyrosine (substrate for p-coumaric acid), but also malonyl-CoA. The current study tested the viability of the module constructs to increase tyrosine production. However, the final goal of this study was to synthesize resveratrol derivatives. Higher yield for resveratrol did not ensure increased production of resveratrol derivatives. Instead, a balance between the upstream and downstream reaction products was critical to determine the final yield. Although the final yield of resveratrol was lower compared to that of the previous studies, it was high enough to synthesize other resveratrol derivatives (See below).
Synthesis of five resveratrol derivatives using a two-culture system
We attempted to synthesize five resveratrol derivatives (Fig. 1). First, we attempted to synthesize pterostilbene in E. coli. One additional gene (ROMT9) was introduced into the strain B-RS3. It was observed that E. coli harboring ROMT9 converted resveratrol to pterostilbene (data not shown). However, the B-RS3 strain harboring ROMT9 synthesized resveratrol instead of pterostilbene. It is likely that the function of ROMT9 was inhibited by some metabolites. Next, a two-culture system, which has been recently implemented for the synthesis of anthocyanin and 4-hydroxy-1-methyl-2-quinolone, was used for the synthesis of resveratrol derivatives [43, 51]. The whole reaction was divided into two steps. In the first step, cells synthesized resveratrol from glucose, while in the second step, they synthesized pterostilbene from resveratrol. The B-RS3 strain was used to synthesize resveratrol (Fig. 3b). The culture filtrate of B-RS3 was heated to destroy the antibiotics. Thereafter, it was mixed with E. coli harboring ROMT9. Culture filtrate from the second mixture was analyzed using HPLC. A new peak with a retention time different from that of resveratrol was observed (Fig. 3c). Mass spectrometry and proton NMR identified the synthesized compound to be pterostilbene. Approximately, 88.2% of the synthesized resveratrol during the first step was converted into pterostilbene in the second reaction.
High performance liquid chromatography (HPLC) analysis of the reaction products after modification of resveratrol. a HPLC profile of standard resveratrol. b HPLC profile of reaction products from B-RS3 strain. P1 and P2 were identified to be p-coumaric acid and resveratrol, respectively. c HPLC profile of reaction products from BL21 strain harboring ROMT9. P3 was determined to be pterostilbene. d HPLC profile of reaction products from BL21 strain harboring SOMT2. P4 was identified to be 4-methoxyresveratrol. e HPLC profile of reaction products from BL21 strain harboring HpaBC. P5 was identified to be piceatannol. f HPLC profile of reaction products from BL21 strain harboring SOMT2 and HpaBC. P6 and P7 were determined to be piceatannol and rhapontigenin, respectively. g HPLC profile of reaction products from BL21 strain harboring ROMT15 and HpaBC. P8 was determined to be isorhapontigenin. All the reaction products were purified and their structures were determined using nuclear magnetic resonance spectroscopy (See Materials and methods)
Four other resveratrol derivatives (4-methoxyresveratrol, piceatannol, rhapontigenin, and isorhapontigenin) were also synthesized using the two-culture system. Each E. coli strain harboring the different modification genes for resveratrol was tested. E. coli strains individually harboring HpaBC and SOMT successfully converted resveratrol from the first cells into piceatannol (Fig. 3e) and 4-methoxyresveratrol (Fig. 3d), respectively. E. coli strains that harbored SOMT2 and HpaBC synthesized rhapontigenin (Fig. 3f), while the ones with ROMT15 and HpaBC synthesized isorhapontigenin (Fig. 3g). The conversion rates for 4-methoxy resveratrol, piceatannol, rhapontigenin, and isorhapontigenin from resveratrol were 72.6%, 71.1%, 22.8%, and 63.5%, respectively. Similar to pterostilbene, structures of the synthesized compounds were determined using proton NMR (Materials and methods).
Anti-inflammatory activity of resveratrol derivatives
Anti-inflammatory effects of resveratrol and five resveratrol derivatives were tested. Relative cell viability and the level of released cytokines were quantitatively measured using protein quantification methods and ELISA after LPS activation with or without resveratrol and resveratrol derivatives (Fig. 4). While resveratrol, 4-methoxyresveratrol, pterostilbene, and isorhapontigenin showed negligible toxicity, rhapontigenin and piceatannol elicited the toxic effect after treatment for 18 h. Cytokine (IL-6 and TNF-α) levels after LPS treatment in the presence of resveratrol derivatives are shown in Fig. 4b, c. Resveratrol and its derivatives successfully reduced the level of both cytokines in RAW264.7 cells. Levels of IL-6 were reduced to 13.8 ± 2.3, 8.3 ± 2.6, 0.5 ± 0.1, 5.9 ± 3.8, 9.6 ± 1.9, and 3.9 ± 0.3 ng/ml upon treatment with resveratrol, 4-methoxyresveratrol, pterostilbene, isorhapontigenin, rhapontigenin, and piceatannol, respectively, compared to the control (80.9 ± 17.8 ng/ml) (Fig. 4b). Previous studies have reported that resveratrol (20–25 μM) reduces the level of IL-6 by 3 ~ 8-folds in LPS-activated RAW264.7 cells (1 μg/mL of LPS) [52, 53]. In the current study, resveratrol (10 μM) induced a fivefold lowering in the IL-6 level, compared to the control; the observation was consistent with the previous studies [52, 53]. TNF-α also showed a significant decrease compared to control upon treatment with resveratrol and its derivatives (Fig. 4c). In particular, pterostilbene exhibited an approximate threefold lowering in the TNF-α release from LPS-activated RAW264.7 cells without any toxicity. Taken together, pterostilbene evidently enhanced the anti-inflammatory effects for macrophages, compared to resveratrol. Considering that methylation of the hydroxyl group at 3` and 5` positions in resveratrol shows increased bioavailability and reduced metabolism, pterostilbene might have a significant anti-inflammatory effect in vivo, compared to resveratrol [54,55,56].
Anti-inflammatory activity of resveratrol derivatives. a Relative cell viability after treatment with lipopolysaccharide (LPS) and resveratrol derivatives. b Level of released interleukin 6 (IL-6) after LPS treatment in the presence of resveratrol derivatives. c Level of tumor necrosis factor α (TNF- α) after LPS treatment in the presence of resveratrol derivatives. Control (Cont), no treatment; 1, resveratrol; 2, 4-methoxyresveratrol; 3, pterostilbene; 4, isorhapontigenin; 5, rhapontigenin; 6, piceatannol. All the compounds were used at a concentration of 10 µM. ns, *, **, and *** indicates not significant, p < 0.05, p < 0.01, and p < 0.001, respectively
In this study resveratrol and five resveratrol derivatives were synthesized and evaluated for their anti-inflammatory activity. Pterostilbene showed the highest yield and the best anti-inflammatory activity. Final yield of pterostilbene can be enhanced if the initial cell concentration, culture time, and/or the copy number of resveratrol modification genes are optimized.
Majority of the biosynthesis studies that focused on the production of a target compound, employed techniques to engineer the host strains. This study not only emphasized on the biosynthesis of diverse derivatives but also attempted to evaluate their biological activity. Synthesis of target compounds with known biological and chemical importance in a biological system is of significant importance. However, at times synthesis of diverse compounds and screening or evaluation of their derivatives might be more critical and prerequisite compared to exploration of strategies to enhance the productivity.
Conclusion
Resveratrol is plant-based phenolic compound and resveratrol derivatives possess anti-inflammatory, anticancer, and antioxidant properties. This study utilized a bacterial host to synthesize resveratrol itself and five resveratrol derivatives. A two-culture system was used for this purpose. The first cell synthesized resveratrol while the second cell comprising resveratrol modification genes synthesized the resveratrol derivatives, namely isorhapontigenin, pterostilbene, 4-methoxyresveratrol, piceatannol, and rhapontigenin. The yield for resveratrol was 89.8 mg/L, while the conversion rate for the resveratrol derivatives was 88.2% to 22.9%. Anti-inflammatory activity of the synthesized resveratrol derivatives was evaluated and pterostilbene was shown the best anti-inflammatory activity. Successful synthesis of resveratrol derivatives with a satisfactory yield using a bacterial host cloned with resveratrol modification genes would indeed serve as a good source for synthesis of resveratrol derivatives.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
References
Kasiotis KM, Pratsinis H, Kletsas D, Haroutounian SA (2013) Resveratrol and related stilbenes: their anti-aging and anti-angiogenic properties. Food Chem Toxicol 61:112–120
Frémont L (2000) Biological effects of resveratrol. Life Sci 66:663–673
Jeandet P, Vannozzi A, Sobarzo-Sanchez E, Uddin Md S, Bru R, Martínez-Marquez A, Clément C, Cordelier S, Manayi A, Nabavi SF, Rasekhian M, Batiha GE-S, Khan H, Morkunas I, Belwal T, Jiang J, Koffaso M, Nabavi SM (2020) Phytostilbenes as agrochemicals: biosynthesis, bioactivity, metabolic engineering and biotechnology. Nat Prod Rep in press
Shen T, Wang X-N, Lou H-X (2009) Natural stilbenes: an overview. Nat Prod Rep 26:916–935
Halls C, Yu O (2008) Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol 26:77–81
Parage C, Tavares R, Réty S, Baltenweck-Guyot R, Poutaraud A, Renault L, Heintz D, Lugan R, Marais GAB, Aubourg S, Hugueney P (2012) Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol 160:407–1419
Morita H, Noguchi H, Schröder J, Abe I (2001) Novel polyketides synthesized with a higher plant stilbene synthase. Eur J Biochem 268:3759–3766
Kodan A, Kuroda H, Sakai F (2002) A stilbene synthase from Japanese red pine (Pinus densiflora): implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc Natl Acad Sci USA 99:3335–3339
Li M, Kildegaard KR, Chen Y, Rodriguez A, Borodina I, Nielsen J (2015) De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab Eng 32:1–11
Li M, Schneider K, Kristensen M, Borodina I, Nielsen J (2016) Engineering yeast for high-level production of stilbenoid antioxidants. Sci Rep 11:36827
Wu J, Zhou P, Zhang X, Dong M (2017) Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J Ind Microbiol Biotechnol 44:1083–1095
Zhao Y, Wu BH, Liu ZN, Qiao J, Zhao GR (2018) Combinatorial optimization of resveratrol production in engineered E. coli. J Agric Food Chem 66:13444–13453
Jeong YJ, An CH, Woo SG, Jeong HJ, Kim YM, Park SJ, Yoon BD, Kim CY (2014) Production of pinostilbene compounds by the expression of resveratrol O-methyltransferase genes in Escherichia coli. Enzyme Microb Technol 54:8–14
Kang SY, Lee JK, Choi O, Kim CY, Jang JH, Hwang BY, Hong YS (2014) Biosynthesis of methylated resveratrol analogs through the construction of an artificial biosynthetic pathway in E. coli. BMC Biotechnol 14:67
Heo KT, Kang SY, Hong YS (2017) De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis. Microb Cell Fact 16:30
Wang S, Zhang S, Xiao A, Rasmussen M, Skidmore C, Zhan J (2015) Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab Eng 29:153–159
Shrestha A, Pandey RP, Pokhrel AR, Dhakal D, Chu LL, Sohng JK (2018) Modular pathway engineering for resveratrol and piceatannol production in engineered Escherichia coli. Appl Microbiol Biotechnol 102:9691–9706
De la Lastra CA, Villegas I (2005) Resveratrol as an anti-inflammatory and anti-aging agent: mechanisms and clinical implications. Mol Nutr Food Res 49:405–430
De Sá CD, Pacheco MT, Frozza RL, Bernardi A (2018) Anti-inflammatory effects of resveratrol: mechanistic insights. Int J Mol Sci 19:1812
Koushki M, Dashatan NA, Meshkani R (2018) Effect of resveratrol supplementation on inflammatory markers: a systematic review and meta-analysis of randomized controlled trials. Clin Ther 40:1180–1192
Shabani M, Sadeghi A, Hosseini H, Teimouri M, Babaei Khorzoughi R, Pasalar P, Meshkani R (2020) Resveratrol alleviates obesity-induced skeletal muscle inflammation via decreasing M1 macrophage polarization and increasing the regulatory T cell population. Sci Rep 10:3791
Remsberg CM, Yáñez JA, Ohgami Y, Vega-Villa KR, Rimando AM, Davies NM (2008) Pharmacometrics of pterostilbene: preclinical pharmacokinetics and metabolism, anticancer, antiinflammatory, antioxidant and analgesic activity. Phytother Res 22:169–179
Paul S, DeCastro AJ, Lee HJ, Smolarek AK, So JY, Simi B, Wang CX, Zhou R, Am R, Suh N (2010) Dietary intake of pterostilbene, a constituent of blueberries, inhibits the beta-catenin/p65 downstream signaling pathway and colon carcinogenesis in rats. Carcinogenesis 31:1272–1278
Roupe KA, Remsberg CM, Yáñez JA, Davies NM (2006) Pharmacometrics of stilbenes: seguing towards the clinic. Curr Clin Pharmacol 1:81–101
Piotrowska H, Kucinska M, Murias M (2002) Biological activity of piceatannol: leaving the shadow of resveratrol. Mutat Res 750:60–82
Seyed MA, Jantan I, Bukhari SNA, Vijayaraghavan K (2016) A comprehensive review on the chemotherapeutic potential of piceatannol for cancer treatment, with mechanistic insights. J Agric Food Chem 64:725–737
Ashikawa K, Majumdar S, Banerjee S, Bharti A, Shishodia S, Aggarwal B (2002) Piceatannol inhibits TNF-induced NF-κB activation and NF-κB-mediated gene expression through suppression of IκBα kinase and p65 phosphorylation. J Immunol 169:6490–6497
Li Y, Yang P, Chang Q, Wang J, Liu J, Lv Y, Wang TTY, Gao B, Zhang Y, Yu LL (2017) Inhibitory effect of piceatannol on TNF-α-mediated inflammation and insulin resistance in 3T3-L1 adipocytes. J Agric Food Chem 65:4634–4641
Yamamoto T, Li Y, Hanafusa Y, Yeh YS, Maruki-Uchida H, Kawakami S, Sai M, Goto T, Ito T, Kawada T (2016) Piceatannol exhibits anti-inflammatory effects on macrophages interacting with adipocytes. Food Sci Nutr 5:76–85
Park EK, Choo MK, Yoon HK, Kim DH (2002) Antithrombotic and antiallergic activities of rhaponticin from Rhei Rhizoma are activated by human intestinal bacteria. Arch Pharm Res 25:528–533
Fan Y (2019) Cardioprotective effect of rhapontigenin in isoproterenol-induced myocardial infarction in a rat model. Pharmacology 103:291–302
Kim JS, Kang CG, Kim SH, Lee EO (2014) Rhapontigenin suppresses cell migration and invasion by inhibiting the PI3K-dependent Rac1 signaling pathway in MDA-MB-231 human breast cancer cells. J Nat Prod 77:1135–1139
Fang Y, Yu Y, Hou Q, Zheng X, Zhang M, Zhang D, Li J, Wu X-R, Huang C (2012) The Chinese herb isolate isorhapontigenin induces apoptosis in human cancer cells by down-regulating overexpression of antiapoptotic protein XIAP. J Biol Chem 287:35234–35243
Li HL, Wang AB, Huang Y, Liu DP, Wei C, Williams GM, Zhang C-N, Liu G, Liu Y-Q, Hao D-L, Hui R-T, Lin M, Liang C (2005) Isorhapontigenin, a new resveratrol analog, attenuates cardiac hypertrophy via blocking signaling transduction pathways. Free Radic Biol Med 38:243–257
Yeo SCM, Fenwick PS, Barnes PJ, Lin HS, Donnelly LE (2017) Isorhapontigenin, a bioavailable dietary polyphenol, suppresses airway epithelial cell inflammation through a corticosteroid-independent mechanism. Br J Pharmacol 174:2043–2059
Lee YJ, Jeon Y, Lee JS, Kim BG, Lee CH, Ahn JH (2007) Enzymatic synthesis of phenolic CoAs using 4-coumarate:coenzyme A ligase (4CL) from rice. Bull Korean Chem Soc 28:365–366
Kim MJ, Kim BG, Ahn JH (2013) Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl Microbiol Biotechnol 97:7195–7204
Lee YJ, Kim BG, Chong Y, Lim Y, Ahn JH (2008) Cation dependent O-methyltransferases from rice. Planta 227:641–647
Cha MN, Kim HJ, Kim BG, Ahn JH (2014) Synthesis of chlorogenic acid and p-coumaroyl shikimates from glucose using engineered Escherichia coli. J Microbiol Biotechnol 24:1109–1117
Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Flavonoid 3’-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67:387–394
Kim DH, Kim BG, Lee Y, Ryu JY, Lim Y, Hur HG, Ahn JH (2005) Regiospecific methylation of naringenin to ponciretin by soybean O-methyltransferase expressed in Escherichia coli. J Biotech 115:155–162
Kim BG, Jung WD, Mok H, Ahn JH (2013) Production of hydroxycinnamoyl-shikimates and chlorogenic acid in Escherichia coli: production of hydroxycinnamic acid conjugates. Microb Cell Fact 12:15
Choo HJ, Ahn JH (2019) Synthesis of three bioactive aromatic compounds by introducing polyketide synthase genes into engineered Escherichia coli. J Agric Food Chem 67:8581–8589
Kim H, Kim SY, Sim GY, Ahn JH (2020) Synthesis of 4-hydroxybenzoic acid derivatives in Escherichia coli. J Agric Food Chem 68:9743–9749
Chung D, Kim SY, Ahn JH (2017) Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli. Sci Rep 7:2578
Yoon JA, Kim BG, Lee WJ, Lim Y, Chong Y, Ahn JH (2012) Production of a novel quercetin glycoside through metabolic engineering of Escherichia coli. Appl Environ Microbiol 78:4256–4262
Chen G, Li KK, Fung CH, Liu CL, Wong HL, Leung PC, Ko CH (2014) Er-Miao-San, a traditional herbal formula containing Rhizoma Atractylodis and Cortex Phellodendri inhibits inflammatory mediators in LPS-stimulated RAW264.7 macrophages through inhibition of NF-kappaB pathway and MAPKs activation. J Ethnopharmacol 154:711–718
Samsonova O, Pfeiffer C, Hellmund M, Merkel OM, Kissel T (2011) Low molecular weight pDMAEMA-block-pHEMA block-copolymers synthesized via RAFT-polymerization: potential non-viral gene delivery agents? Polymer 3:693–718
Park JY, Lim J-H, Ahn J-H, Kim B-G (2021) Biosynthesis of resveratrol using metabolically engineered Escherichia coli. Appl Biol Chem 64:20
Choi GS, Choo HJ, Kim BG, Ahn JH (2020) Synthesis of acridone derivatives via heterologous expression of a plant type III polyketide synthase in Escherichia coli. Microb Cell Fact 19:73
Jones JA, Vernacchio VR, Collins SM, Shirke AN, Xiu Y, Englaender JA, Cress BF, McCutcheon CC, Linhardt RJ, Gross RA, Koffas MA (2017) Complete biosynthesis of anthocyanins using E. coli polycultures. MBio 8:e00621-e717
Ma C, Wang Y, Dong L, Li M, Cai W (2015) Anti-inflammatory effect of resveratrol through the suppression of NF-kappaB and JAK/STAT signaling pathways. Acta Biochim Biophys Sin 47:207–213
Tong W, Chen X, Song X, Chen Y, Jia R, Zou Y, Li L, Yin L, He C, Liang X, Ye G, Lv C, Lin J, Yin Z (2020) Resveratrol inhibits LPS-induced inflammation through suppressing the signaling cascades of TLR4-NF-κB/MAPKs/IRF3. Exp Ther Med 19:1824–1834
Arbo BD, André-Miral C, Nasre-Nasser RG, Schimith LE, Santos MG, Costa-Silva D, Muccillo-Baisch AL, Hort MA (2020) Resveratrol derivatives as potential treatments for Alzheimer’s and Parkinson’s disease. Front Aging Neurosci 12:103
Dvorakova M, Landa P (2017) Anti-inflammatory activity of natural stilbenoids: a review. Pharmacol Res 124:126–145
Freyssin A, Page G, Fauconneau B, Rioux Bilan A (2020) Natural stilbenes effects in animal models of Alzheimer’s disease. Neural Regen Res 15:843–849
Acknowledgements
The present study was supported by grants from the National Research Foundation (NRF298 2019R1A2C1002714) funded by the Ministry of Education, Science and Technology Republic of Korea.
Funding
Funding was received from the National Research Foundation (NRF298 2019R1A2C1002714) funded by the Ministry of Education, Science and Technology 299 Republic of Korea.
Author information
Authors and Affiliations
Contributions
HM and JHA designed the experiments. YC, HLL, JS, YL, BGK and JHA performed the experiments and analyzed the data. YC, HLL, BGK, HM and JHA wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chong, Y., Lee, H.L., Song, J. et al. Biosynthesis of resveratrol derivatives and evaluation of their anti-inflammatory activity. Appl Biol Chem 64, 33 (2021). https://doi.org/10.1186/s13765-021-00607-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-021-00607-4