Abstract
Background
The emergence of carbapenemase-producing (CP) Citrobacter freundii poses a significant threat to public health, especially in high-risk populations. In this study, whole genome sequencing was used to characterize the carbapenem resistance mechanism of three C. freundii clinical isolates recovered from fecal samples of patients with acute leukemia (AL) from Spain.
Materials and methods
Twelve fecal samples, collected between 2013 and 2015 from 9 patients with AL, were screened for the presence of CP strains by selecting them on MacConkey agar supplemented with ertapenem (0.5 mg/L). Bacteria were identified by MALDI-TOF mass spectrometry and were phenotypically characterized. Whole genome sequencing of C. freundii isolates was performed using the MinION and MiSeq Illumina sequencers. Bioinformatic analysis was performed in order to identify the molecular support of carbapenem resistance and to study the genetic environment of carbapenem resistance encoding genes.
Results
Three carbapenem-resistant C. freundii strains (imipenem MIC≥32 mg/L) corresponding to three different AL patients were isolated. Positive modified Carba NP test results suggested carbapenemase production. The genomes of each C. freundii tested were assembled into a single chromosomal contig and plasmids contig. In all the strains, the carbapenem resistance was due to the coproduction of OXA-48 and VIM-1 enzymes encoded by genes located on chromosome and on an IncHI2 plasmid, respectively. According to the MLST and the SNPs analysis, all strains belonged to the same clone ST169.
Conclusion
We report in our study, the intestinal carrying of C. freundii clone ST169 coproducing OXA-48 and VIM-1 identified in leukemic patients.
Similar content being viewed by others
Introduction
In patients with acute leukemia (AL), long duration and repetitive chemotherapy as well as antimicrobial therapy is believed to contribute to occurrence of infections due to multi-drug resistant (MDR) bacteria in this high-risk group [1, 2]. In patients with leukemia, because of therapy of their diseases, bacterial infection with MDR Gram-negative bacteria is a real problem that could be associated with a high rate of mortality and morbidity [2,3,4].
Citrobacter freundii is a gram-negative bacterium which is rarely the causative agent of infections but it has been associated with different infections including respiratory, urinary, gastrointestinal and bloodstream infections [5, 6]. The emergence of MDR C. freundii, especially those carbapenemase producing strains, poses a significant threat to public health worldwide, especially in immunocompromised patients such as leukemia patients, which are mostly dependent on antibiotics [5, 6].
Since the development of new generation sequencing technologies, the access to the full genetic bacterial repertoire has become easier and allow a better understanding the emergence of antibiotic resistance genes on a global scale [7, 8].
In this study, we applied the whole-genome sequencing to characterize the antibiotic resistance mechanisms of three carbapenem-resistant C. freundii clinical isolates recovered from fecal samples of patients with acute leukemia from Spain.
Materials and methods
Study design, bacterial isolates and antimicrobial susceptibility test
The Study design was described in our previous publication [9]. A subset of twelve fecal samples, collected between 2013 and 2015 from 9 patients with AL, were screened for the presence of carbapenemase producing (CP) strains by selecting them on MacConkey agar supplemented with ertapenem (0.5 mg/L) [10]. Cultivated bacteria were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Microflex, Bruker Daltonics, Bremen, Germany). The resistance phenotype of the isolates was evaluated by testing their susceptibly against sixteen antibiotics on Mueller Hinton agar using disk diffusion methodology according to the European Committee on Antimicrobial Susceptibility testing (EUCAST) guidelines (http://www.eucast.org). The minimum inhibitory concentration (MIC) of imipenem was determined using Etest method (AB Biodisk, Sweden), the results were interpreted according to the EUCAST breakpoint. The modified Carba NP test method was used to determine a possible carbapenemase production [11].
Genetic and genomic characterization
Real-time and standard PCR were performed to screen for the presence of carbapenem resistance genes, including blaOXA-48, blaKPC, blaNDM and blaVIM [12]. Whole genome sequencing of the CP strains was performed using the MinION (Oxford Nanopore Technologies Inc., UK) and the MiSeq (Illumina Inc., San Diego, CA, USA) technologies in order to determine the carbapenemase genes variants, the genetic environment, and the genetic support of these genes. The long-read sequencing data generated by Nanopore and short-read data produced by Illumina sequencing were assembled using SPAdes genome assembler [13]. ARG-ANNOT database available on the ABRicate pipeline and Prokka software were used to identify the antibiotic resistances genes and to annotate genomes, respectively [14, 15]. Genetic environment has been reconstituted by comparing the sequence of genes surrounding the carbapenemase gene against the NCBI database, using blastX parameter.
Clonal relationship
SNPs analysis (available at https://cge.cbs.dtu.dk/services/CSIPhylogeny/) was conducted to study the genomic difference between the three strains and to determine the possible clonal relationship. In order to determine the sequence type (ST) of isolated strains, Multi Locus Sequence Typing (MLST) analysis was performed in silico using the MLST database (available at https://cge.cbs.dtu.dk/services/MLST/).
Conjugation experiments and plasmid analysis
Conjugation was conducted on the three C. freundii isolates using azide-resistant Escherichia coli J53 as a recipient, as previously described [16]. The transconjugants were selected on Luria Bertani (LB) agar (Beckton Dickinson, Le Pont de Claix, France) supplemented with sodium azide (120 μg/ml) and Ertapenem (2 μg/ml). Plasmids analysis was performed in silico. Plasmid incompatibility type was identified using PlasmidFinder database (available at https://cge.cbs.dtu.dk/services/PlasmidFinder/) and Jspecies Web Server was used to calculate the extent of identity between the plasmids [17].
Nucleotide sequence accession number
The shotgun whole genome sequence of the three C. freundii strains and complete sequence of plasmids have been deposited in NCBI GenBank (GenBank accession number CP038653, CP038654, CP038655, CP038656, CP038657, CP038658, CP038659 and CP038660).
Results
Bacterial strains and microbiological tests
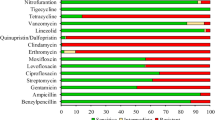
Three C. freundii strains (C. freundii_154, C. freundii_565 and C. freundii_680) were isolated on selective media from fecal samples of three different leukemic patients (Patient-1, Patient-2 and Patient-3) aged 49, 40, 51, respectively (Table 1). All patients received ciprofloxacin prior sampling and only one (Patient-2) received also meropenem (Table 1). Before samples collection, Patient-1 and Patient-2 received an allogenic transplantation, whereas Patient-3 received a high-intensity chemotherapy. The three strains were resistant to most antibiotics tested (Table 2), including carbapenems with imipenem MIC ≥32 μg/ml. All the strains remained susceptible to doxycycline, colistin, fosfomycin and nitrofurantoin. Positive modified Carba NP test results suggested carbapenemase production.
Genetic, genomic and molecular epidemiology analysis
The genome size of CP C. freundii strains obtained after assembly ranged from 5′443’022 and 5′471’065 bp (including chromosome and plasmids for each strains) (Table 2). According to PCR results and genome analysis, carbapenem resistance in these strains was due to the co-production of OXA-48 and VIM-1 carbapenemase enzymes. Resistome analysis showed the presence of genes encoding for resistance to β-lactams, aminoglycosides, quinolones, sulfonamides, trimethoprim and chloramphenicol antibiotics families (Table 2). The gene encoding OXA-48 enzyme was located on the chromosome in the three strains tested and surrounded by the almost similar structures that compose the Tn1999.2 transposon (ΔTn1999/IS1R-blaOXA-48-LysR-orf-ISL3-like) (Fig. 1.a). Unlike the blaOXA-48 gene, blaVIM-1 gene was located in a IncHI-2 plasmid in all the strains. This gene was located in a class 1 integron that contains a new cassette array (intI1–blaVIM-1-aac6-Ib-cr-aadA1-qqcEΔ1/sul1-ΔtniB3-tniA) (Fig. 1b). According to the MLST analysis, all strains belonged to the same sequence type, ST169. The SNPs analysis found between 10 and 19 SNPs on average between the three isolates suggesting that these ST169 strains belonged to the same clone.
a Schematic representation of the genetic environment of the blaOXA-48 gene located on chromosome and its comparison with the Tn1999 transposon (JN626286) and its variant Tn1999.2 (JN714122) and Tn1999.3 (HE617182). b Schematic representation of genes surrounded the blaVIM-1 gene and the comparison of this genetic environment with that one identified in the Tn402-type C
Plasmid conjugation
Conjugation experiments failed for the three strains tested, whereas, in silico plasmids analysis showed the presence of different protein implicated in conjugal transfer and pilus formation but the absences of the plasmid transfer origin (oriT). The comparison of the average nucleotide identity between the three plasmids harboring blaVIM-1 gene showed that plasmid p154_1 shared 99.99% of similarity with the plasmid p565_1, whereas p680_1 shared 99.94% of similarity with the two other plasmids.
Discussion
Carbapenemase production in C. freundii is poorly documented, only a few studies reported the expression of such enzymes in this species [6, 18,19,20]. The coproduction of carbapenemase enzymes was already described in some Enterobacteriaceae species such as Klebsiella pneumoniae (KPC-2 + VIM-2 or NDM-1 + VIM-1) [19, 21, 22] and Enterobacter cloacae (NDM-1 + VIM-1) [19] as well as in C. freundii (KPC-2 + NDM-1, NDM-1 + VIM-1) [19, 20]. In our study, this situation was observed in three carbapenem resistant C. freundii strains detected in fecal samples of three AL patients where, interestingly, two of them didn’t received carbapenem as antimicrobial therapy. This suggest that the carbapenem resistance in this context may not be due to a selection pressure with this antibiotic family but it could have been selected by the use of other antibiotic families or by the presence of a carbapenem-resistant clone in the hematological ward, which would explain this current situation.
In Spain, OXA-48 and VIM-1 enzymes are the most prevalent carbapenemase enzymes reported especially in E. coli, E. cloacae and K. pneumoniae [18]. The coproduction of these two carbapenemase enzymes by C. freundii species was reported in only three studies over the world, and only one reported this detection in hematological malignancies patients [18, 19, 23]. During an unrestricted and non-mandatory national Spanish Antibiotic Resistance Surveillance Programme, undertaken between 2013 and 2015, it has been noted a progressive increase in the rate of Citrobacter spp. Carbapenemase-producers, including C. freundii species, in Spanish hospital from 1.3% in 2013 to 1.5% in 2015 [18].
The gene encoding OXA-48 enzyme was mainly related to the Tn1999 transposon and to its variants [24]. Our study showed that the blaOXA-48 gene was located on the chromosome in all strains tested and that its genetic environment was almost identical to that described in Tn1999.2 variant (Table 2, Fig. 1a) [24]. Indeed, the only difference resides downstream of the blaOXA-48 gene, where this gene was flanked by an ISL3-like in our three C. freundii instead of IS1999 described in Tn1999.2 variant (Fig. 1a).
blaVIM-1 gene was widely detected in different class 1 integrons such as In110 or In113 [25]. In the study conducted by Arana et al in Spain, blaVIM-1 identified in their C. freundii strains was located on class 1 integrons which include other antibiotics resistant genes such as aacA4, dfrB1, aadA1, and catB2genes [18]. Our study also reports the localization of the blaVIM-1 gene in a class 1 integrons, which contains a new gene cassette, composed of the blaVIM-1, aac6-Ib-cr (conferring resistance to both aminoglycosides and quinolones), aadA1 (conferring resistance to aminoglycosides) as well as the classic sulI gene (Fig. 1b). The structure of the integrons class 1 type identified in our strains looks like the defective Tn402 transposon (type C) carrying the tni module, ∆tniB and tniA, reported in the literature (Fig. 1b) [26].
In our study, the plasmid carrying the blaVIM-1 gene identified cannot conjugate, thus excluding the possibility of plasmid dissemination between patients. Moreover, MLST and SNPs analysis showed that the three C. freundii strains belonged to the same ST169 clone, which leads us to hypothesize a possible clonal spread of carbapenem-resistant strains in the hematology department.
In this present study, the three C. freundii coproducing OXA-48 and VIM-1 carbapenemase enzymes were isolated in a context of digestive carrying and not infectious. It has been shown that in hematological patients, colonization of the digestive tract by carbapenem-resistant bacteria constitutes a risk in the development of infections with these bacteria [27,28,29]. Despite the fact that our isolates exhibited a high resistance profile, some antibiotics remained active on these bacteria such as doxycycline, colistin, fosfomycin or nitrofurantoin.
Conclusion
This study reports the clonal spread of C. freundii ST169 exhibiting a rare phenotype of co-production of two carbapenemases, namely OXA-48 and VIM-1 enzymes, detected in the digestive tract of patients with acute leukemia. In our opinion, a systematic screening of digestive carriage of antibiotics resistant bacteria would be a great solution to prevent the occurrence of infections due to such bacteria and to control the spread of antibiotic resistance genes, especially within high risk populations.
Availability of data and materials
Not applicable.
Abbreviations
- AL:
-
Acute leukemia
- ARG-ANNOT:
-
Antibiotic resistance gene-annotation
- CP:
-
Carbapenemase producing
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- LB:
-
Lucia Bertani
- MALDI-TOF MS:
-
Matrix-assisted laser desorption and ionization time-of-flight mass spectrometry
- MDR:
-
Multi drug resistant
- MIC:
-
Minimum inhibitory concentration
- MLST:
-
Multi-locus sequence typing
- NCBI:
-
National Center for Biotechnology Information
- PCR:
-
Polymerase chain reaction
- SNPs:
-
Single nucleotide polymorphisms
- ST:
-
Sequence type
References
Alp S, Akova M. Antibacterial resistance in patients with hematopoietic stem cell transplantation. Mediterr J Hematol Infect Dis. 2017;9:e2017002.
Trubiano JA, Worth LJ, Thursky KA, Slavin MA. The prevention and management of infections due to multidrug resistant organisms in haematology patients. Br J Clin Pharmacol. 2015;79:195–207.
Baker TM, Satlin MJ. The growing threat of multidrug-resistant gram-negative infections in patients with hematologic malignancies. Leuk Lymphoma. 2017;57:2245–58.
Blennow O, Ljungman P. The challenge of antibiotic resistance in haematology patients. Br J Haematol. 2016;172:497–511.
Yang L, Peihan L, Beibei L, Xiaofeng H, Jinhui L, Jing X, et al. Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci Rep. 2018;8:1–7. Available from:. https://doi.org/10.1038/s41598-018-28879-9.
Ouyang J, Xiong Z, Yang B, Liu Z, Sun F, Zhou D, et al. Comparative genomics of five different resistance plasmids coexisting in a clinical multi-drug resistant Citrobacter freundii isolate. Infect Drug Resist. 2018;11:1447–60.
Bakour S, Sankar SA, Rathored J, Biagini P, Didier R, Fournier P-E. Identification of virulence factors and antibiotic resistance markers using bacterial genomics. Future Microbiol. 2016;11:455–66.
Hadjadj L, Baron SA, Diene SM, Rolain J-M. How to discover new antibiotic resistance genes? Expert Rev Mol Diagn. 2019;19:1–14. https://doi.org/10.1080/14737159.2019.1592678 Taylor & Francis. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30895843%0A https://www.tandfonline.com/.
Lalaoui R, Djukovic A, Bakour S, Sanz J, Gonzalez-Barbera EM, Salavert M, et al. Detection of plasmid-mediated colistin resistance, mcr-1 gene, in Escherichia coli isolated from high-risk patients with acute leukemia in Spain. J Infect Chemother. 2019;25:605–609.
Bachiri T, Bakour S, Lalaoui R, Belkebla N, Allouache M, Rolain JM, et al. Occurrence of carbapenemase-producing enterobacteriaceae isolates in the wildlife: first report of OXA-48 in wild boars in Algeria. Microb Drug Resist. 2017;24:337–45.
Bakour S, Garcia V, Loucif L, Brunel J-M, Gharout-Sait A, Touati A, et al. Rapid identification of carbapenemase-producing Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii using a modified Carba NP test. New Microbes New Infect. 2015;7:89–93 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26442150. [cited 2019 May 22].
Mellouk FZ, Bakour S, Meradji S, Al-Bayssari C, Bentakouk MC, Zouyed F, et al. First Detection of VIM-4-Producing Pseudomonas aeruginosa and OXA-48-Producing Klebsiella pneumoniae in Northeastern (Annaba, Skikda) Algeria. Microb Drug Resist. 2017;23:335–44 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27314660. [cited 2019 May 22].
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 2012;19:455–77 Available from: http://www.ncbi.nlm.nih.gov/pubmed/22506599. [cited 2019 mar 22].
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob Agents Chemother. 2014;58:212–20 Available from: http://www.ncbi.nlm.nih.gov/pubmed/24145532. [cited 2019 Mar 22].
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9 Available from: http://www.ncbi.nlm.nih.gov/pubmed/24642063. [cited 2019 Mar 22].
Lalaoui R, Bakour S, Livnat K, Assous M, Diene S, Rolain J. Spread of Carbapenem and Colistin-resistant Klebsiella pneumoniae ST512 clinical isolates in Israel: a cause for vigilance. Microb Drug Resist. 2019;25:63–71.
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. Available from: [cited 2019 Mar 22]. https://doi.org/10.1093/bioinformatics/btv681.
Arana DM, Ortega A, González-Barberá E, Lara N, Bautista V, Gómez-Ruíz D, et al. Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J Antimicrob Chemother. 2017;72:3283–7.
Bedenić B, Sardelić S, Luxner J, Bošnjak Z, Varda-Brkić D, Lukić-Grlić A, et al. Molecular characterization of class b carbapenemases in advanced stage of dissemination and emergence of class d carbapenemases in Enterobacteriaceae from Croatia. Infect Genet Evol. 2016;43:74–82 Available from: https://linkinghub.elsevier.com/retrieve/pii/S1567134816301836. [cited 2019 Mar 22].
Feng J, Qiu Y, Yin Z, Chen W, Yang H, Yang W, et al. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob Chemother. 2015;70:2987–91. Available from: [cited 2019 Mar 22]. https://doi.org/10.1093/jac/dkv232.
Papagiannitsis CC, Malli E, Florou Z, Sarrou S, Hrabak J, Mantzarlis K, et al. Emergence of sequence type 11 Klebsiella pneumoniae coproducing NDM-1 and VIM-1 metallo-β-lactamases in a Greek hospital. Diagn Microbiol Infect Dis. 2017;87:295–7 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0732889316304266. [cited 2019 Mar 22].
Falco A, Ramos Y, Franco E, Guzmán A, Takiff H. A cluster of KPC-2 and VIM-2-producing Klebsiella pneumoniae ST833 isolates from the pediatric service of a Venezuelan Hospital. BMC Infect Dis. 2016;16:595 Available from: http://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-016-1927-y. [cited 2019 Mar 22].
Jayol A, Poirel L, Dortet L, Nordmann P. National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Euro Surveill. 2016;21:30339 Available from: http://www.eurosurveillance.org/ViewArticle.aspx? ArticleId=22579. [cited 2019 Mar 22].
Machuca J, López-Cerero L, Fernández-Cuenca F, Mora-Navas L, Mediavilla-Gradolph C, López-Rodríguez I, et al. OXA-48-like-producing Klebsiella pneumoniae in southern Spain in 2014–2015. Antimicrob Agents Chemother. 2019;63:e01396-18.
Porres-Osante N, Azcona-Gutiérrez JM, Rojo-Bezares B, Undabeitia E, Torres C, Sáenz Y. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J Antimicrob Chemother. 2014;69:1792–5.
Tato M, Coque TM, Baquero F, Cantón R. Dispersal of carbapenemase blaVIM-1 gene associated with different Tn402 variants, mercury transposons, and conjugative plasmids in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:320–7.
Jaiswal SR, Gupta S, Kumar RS, Sherawat A, Rajoreya A, Dash SK, et al. Gut colonization with carbapenem-resistant enterobacteriaceae adversely impacts the outcome in patients with hematological malignancies: results of a prospective surveillance study. Mediterr J Hematol Infect Dis. 2018;10:1–8.
Fanci R, Sica S, Cattaneo C, Fianchi L, Busca A, Trecarichi EM, et al. Bloodstream infections caused by Klebsiella pneumoniae in onco-hematological patients: clinical impact of carbapenem resistance in a multicentre prospective survey. Am J Hematol. 2016;91:1076–81.
Andria N, Henig O, Kotler O, Domchenko A, Oren I, Zuckerman T, et al. Mortality burden related to infection with carbapenem-resistant gram-negative bacteria among haematological cancer patients: a retrospective cohort study. J Antimicrob Chemother. 2015;70:3146–53.
Acknowledgements
The authors thank Pr. Karina Xavier, Pr. Bernhard Kuster and Pr. Laurent Debrauwer for their contribution. They also thank CookieTrad for proofreading the text.
Funding
This work was supported by the ANR FloraStopInfectMRE project, InfecERA-ERANET-Acciones complementarias (PCIN-2015-094) to CU and by IHU Méditerranée Infection, Marseille, France and by the French Government under the «Investissements d’avenir» (Investments for the Future) proGram managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU- 03).
This work was supported by Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
Author information
Authors and Affiliations
Contributions
RL performed experiments, analyzed the data and wrote the manuscript. AD obtained all the samples and clinical data from the patients. SB analyzed the data and contributed to the preparation of the manuscript. LH performed the genome sequencing. JS, MS, JLL-H and MAS contributed to the obtention of all the clinical samples and clinical data from the patients. CU contributed to the design of the study. J-MR designed the study and helped draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All subjects gave their informed consent for inclusion before they participated in the study. The study was approved on the 1st of July 2013 by the Ethics Committee of CEIC Dirección General de Salud Pública y Centro Superior de Investigación en Salud Pública (20130515/08).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lalaoui, R., Djukovic, A., Bakour, S. et al. Genomic characterization of Citrobacter freundii strains coproducing OXA-48 and VIM-1 carbapenemase enzymes isolated in leukemic patient in Spain. Antimicrob Resist Infect Control 8, 167 (2019). https://doi.org/10.1186/s13756-019-0630-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-019-0630-3