Abstract
Background
Antibiotic misuse in food-producing animals is potentially associated with human acquisition of multidrug-resistant (MDR; resistance to ≥ 3 drug classes) bacteria via the food chain. We aimed to determine if MDR Gram-negative (GNB) organisms are present in fresh Australian chicken and pork products.
Methods
We sampled raw, chicken drumsticks (CD) and pork ribs (PR) from 30 local supermarkets/butchers across Melbourne on two occasions. Specimens were sub-cultured onto selective media for third-generation cephalosporin-resistant (3GCR) GNBs, with species identification and antibiotic susceptibility determined for all unique colonies. Isolates were assessed by PCR for SHV, TEM, CTX-M, AmpC and carbapenemase genes (encoding IMP, VIM, KPC, OXA-48, NDM).
Results
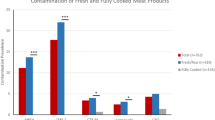
From 120 specimens (60 CD, 60 PR), 112 (93%) grew a 3GCR-GNB (n = 164 isolates; 86 CD, 78 PR); common species were Acinetobacter baumannii (37%), Pseudomonas aeruginosa (13%) and Serratia fonticola (12%), but only one E. coli isolate. Fifty-nine (36%) had evidence of 3GCR alone, 93/163 (57%) displayed 3GCR plus resistance to one additional antibiotic class, and 9/163 (6%) were 3GCR plus resistance to two additional classes. Of 158 DNA specimens, all were negative for ESBL/carbapenemase genes, except 23 (15%) which were positive for AmpC, with 22/23 considered to be inherently chromosomal, but the sole E. coli isolate contained a plasmid-mediated CMY-2 AmpC.
Conclusions
We found low rates of MDR-GNBs in Australian chicken and pork meat, but potential 3GCR-GNBs are common (93% specimens). Testing programs that only assess for E. coli are likely to severely underestimate the diversity of 3GCR organisms in fresh meat.
Similar content being viewed by others
Background
The emergence of multi-drug resistant (MDR) bacteria is a major health problem that has been likened in its global future impact on human health to that of terrorism [1, 2]. Widespread inappropriate use of antimicrobials in food production (especially meat/seafood, some fruit) has been linked to environmental contamination with MDR pathogens and outbreaks of MDR infections in humans, but direct cause-and-effect has often been difficult to confirm, despite the strength of the observed associations [3–8]. Most food testing programs for antimicrobial resistance (AMR) have focused on specific organisms (e.g. E. coli, Salmonella spp., Listeria spp.), assuming direct food-to-human pathogen transfer, rather than considering resistant gene transfer between bacterial species [6, 8, 9]. Furthermore, the optimum site of specimen collection (e.g. on-farm animal, manure, abattoir, point-of-sale supermarket products) has been debated [9–13]. Although Australia has reasonably strict regulations regarding antimicrobial use in agriculture [13, 14], use of some agents for prophylaxis and treatment (e.g. trimethoprim-sulfamethoxazole, some beta-lactams and macrolides) is common in some food sectors [2, 13, 15, 16], such that this may have some implications for acquisition by consumers of multi-resistant pathogens via food consumption [6, 7].
Hence, we aimed to assess the rates of contamination with potential extended-spectrum beta-lactamase (ESBL)-producing Gram-negative organisms (without restricting to specific species) in Australian-produced chicken and pork meat. To best identify any potential risk to the consumer and to be certain that the meat was produced in Australia, we purchased chicken drumsticks and pork ribs at local fresh food outlets, since national legislation requires that bone-containing meat products must be Australian-produced (by conventional or organic production), whereas de-boned meats (e.g. bacon) can be imported into Australia [17].
Methods
Study design
This was a prospective cross-sectional survey undertaken during a four-month period from March to June 2014 in the eastern suburbs of Melbourne, Australia. We identified ten regions within the medical catchment area of the Austin Hospital and sampled from 2 to 4 retailers within each region (see Additional file 1: Figure S1 for locations). We tested raw, skin-covered chicken drumsticks (CD) and pork spare ribs (PR), each weighing approximately 150 g, from a total of 30 meat retailers (26 supermarkets, 4 butchers shops). Samples were purchased from each site on two occasions (approximately one month between each sample). Each sample from a supermarket was derived from a pre-packaged container with multiple CD or PR specimens, while each sample from a butcher was selected for purchase individually and was not pre-packaged.
Specimen handling, culture and susceptibility
Similar to methods previously described [18, 19], each specimen was placed individually into separate zip lock bags (22 × 22 cm, Hercules, Australia), to which 100 mL of buffered peptone water (Thermofisher Scientific, Australia) was added and the specimen was massaged manually for 2 min. Of the subsequent rinsate, 50 mL was added in a sterile manner to 50 mL of double-strength tryptone soya broth (TSB; Thermofisher Scientific, Australia) which was incubated for 24 h at 37 °C. From this broth, 100 μL was inoculated into 10 mL of TSB containing ceftriaxone (0.25 mg/L) and vancomycin (8 mg/L), and incubated (37 °C, 24 h) before 10 μL was inoculated and spread onto ChromID ESBL agar (BioMérieux, France) and incubated for 48 h at 37 °C [18, 19]. All unique colonial morphologies on this selective medium were purity-plated onto Columbia horse blood agar/MacConkey agar (HBA/MAC; Thermofisher Scientific, Australia) and then subcultured onto Columbia HBA (Thermo Scientific, Australia) and incubated (37 °C, 24 h) before being identified using MALDI-TOF MS (BioMérieux, France) and tested for antibiotic susceptibility by Vitek2® (BioMérieux, France) using CLSI clinical breakpoint criteria. For those species and antibiotics where there were no defined criteria (e.g. Pseudomonas spp., Stenotrophomonas spp.), the Vitek-derived MIC value was compared to the relevant EUCAST distribution to categorise (for the purpose of this study) the presence of resistance (either intrinsic or acquired) [20–23]. If MALDI-TOF MS was unable to confidently identify (< 90% match) the organism after three attempts, Vitek2® was used for identification.
Isolates which grew on ChromID ESBL agar and displayed phenotypic resistance by Vitek2 to third-generation cephalosporins (ceftriaxone; 3GCR) were considered to be potential ESBL-producers or intrinsically 3GCR [24–26] and were classified according to the number of antibiotic classes to which they were resistant – including third-generation cephalosporins, carbapenems (meropenem), aminoglycosides (gentamicin), fluoroquinolones (ciprofloxacin) and anti-folates (trimethoprim-sulfamethoxazole). Similar to previously, multi-drug-resistance (MDR) was defined as resistance to ≥3 classes of antibiotics [27].
Molecular assessment for beta-lactamase genes
DNA was extracted from all potential ESBL-producing isolates using previously described methods (DNeasy Blood and Tissue kit, Qiagen, USA), then screened for the presence of the blaTEM, and blaSHV genes using a real-time polymerase chain reaction (PCR) platform (LC-480) and published primers [28, 29]. A multiplex real-time TaqMan PCR was used to detect CTX-M-type genes (groups 1, 2, 9, 8, 25) [30]. Strains were probed for plasmid-borne AmpC enzymes using the method described by Pérez-Pérez and Hanson (including blaACC-like, blaDHA-like, blaCIT/CMY-like, blaMOX-like, blaFOX-like, blaMIR/ACT-like; [31]) and subjected to molecular tests for MBL (blaVIM, blaIMP, and blaNDM), blaKPC, and blaOXA-48-like genes using real-time PCR [32, 33]. Isolates suspected of containing transferable ESBL or MBL genes underwent whole genome sequencing whereby unique dual indexed libraries were prepared from genomic DNA using the Nextera XT DNA sample preparation kit (Illumina). Libraries were sequenced on the Illumina NextSeq 500 with 150-cycle paired end chemistry as described by the manufacturer’s protocols and sequences were accessed for known resistance genes using KmerResistance 2.2 [34].
Data analysis and statistics
The rates of contamination with potential ESBL-producing Gram-negative organisms were assessed according to specimen type (CD, PR), the geographic site of specimen purchase and the type of meat outlet (supermarket vs butcher). Similar rates were reported for PCR-confirmed ESBL isolates and those where the ESBL was likely to be plasmid-mediated. Comparisons between rates for CD and PR were undertaken using Chi-square.
Results
Of a total of 120 meat specimens (60 CD, mean ± SD weight: 155.4 ± 26.5 [range 78.5–223.9] grams; 60 PR, 160.5 ± 48.9 [range: 91.5–355.1] grams) that were assessed from 30 retailers (see locations in Additional file 1: Figure S1), 112 (56 CD, 93%; 56 PR; 93%) were contaminated with a total of 164 (86 CD; 78 PR) 3GCR (i.e. potential ESBL-producing) isolates (Table 1). Among these isolates, 59 (36%; 26 CD, 33 PR) displayed phenotypic evidence of 3GCR alone, 96 (59%; 54 CD, 42 PR) were 3GCR plus were also resistant to either anti-folates, aminoglycosides or carbapenems and 9 isolates (5.5%; 6 CD, 3 PR; 9 specimens; 5 Pseudomonas aeruginosa, 2 Pseudomonas spp., 1 Bordetella trematum, 1 Chryseobacterium gleum) were MDR with evidence of being 3GCR plus resistance to two other antibiotic classes. Resistance to anti-folates was most common (n = 91 [55%] isolates, 49 CD, 42 PR, Table 1; 82 [68.3%] specimens). The four most common 3GCR species identified were Acinetobacter baumannii complex (n = 59), Pseudomonas aeruginosa (n = 22), Serratia fonticola (n = 19) and Hafnia alvei (n = 15). Only one E. coli isolate was identified – this was in a CD specimen.
Among the 164 isolates, 158 had DNA available for PCR analysis. Beta-lactamase genes were identified in 23 (15%) isolates (7CD, 14PR [2 PR each had two isolates], p = 0.15; 17.5% specimens). All were AmpC, with 22/23 considered to be inherently chromosomally located (ACC, n = 12 [H. alvei, 10; S. fonticola, 2]; CMY-like n = 7 [C. freundii, 6; C. youngae/freundii, 1]; FOX, n = 1 [A. sobria]; MIR-like/ACT-like, n = 2; [E. cloacae complex]), while the sole E. coli isolate contained a CMY-like AmpC gene that was likely to be plasmid-mediated and was subsequently shown on whole genome sequencing to be a CMY-2 (see Table 1). All DNA samples were PCR-negative for other ESBL genes (including SHV, TEM, CTX-M) and all carbapenemase encoding gene families (including IMP, VIM, KPC, OXA-48-like and NDM).
Among the 30 food outlets, there were four supermarket chains (two large [n = 10 and 11 stores sampled]; two smaller [n = 2 and 3 stores] and 4 separate (unlinked) butcher shops. Overall, there were no differences in rates of contamination between supermarkets and unlinked butchers shops. All supermarkets and butcher shops had at least one CD or PR specimen that grew a potential ESBL-producing isolate, at some time. Only 8 specimens were culture-negative (4 CD, 4 PR; one supermarket site had both its PR specimens culture-negative). The numbers of 3GCR isolates per specimen were as follows: single isolate in 63 specimens; two isolates in 44 specimens; 3 isolates in 3 specimens, and one specimen contained 4 potential ESBL-producing isolates. Interestingly, it was this latter specimen (which was collected from a butcher’s shop) that grew the CMY-2-containing E. coli, along with an A. baumannii, S. fonticola and an E. cloacae complex isolate – although none of these latter 3 isolates contained any definable ESBL genes (Table 1).
Discussion
This study of Australian chicken and pork is notable for a number of reasons. Firstly, we assessed for a broad range of Gram-negative organisms, not simply the traditional species of E. coli or Salmonella spp. [6, 8–10]. Taking this approach, we identified that 93% of specimens appeared to be contaminated with a wide variety of 3GCR species, including particularly Acinetobacter baumannii complex, Pseudomonas aeruginosa, Serratia fonticola and Hafnia alvei. We were surprised by the relatively high rates of these potential pathogens and initially speculated that perhaps they were due to a point-source within certain supermarkets or butcher shops, such as has been reported in one outbreak of multidrug-resistant K. pneumoniae [35]. However, they were identified from both CD and PR products purchased from a wide variety of food outlets which had no common supply chain. Notably, only one E. coli isolate was identified – so testing programs which only assess for this species would have reported a much lower rate of potential contamination.
Secondly, our results highlight the importance of not relying solely on selective media such as ChromID ESBL agar in such programs, but instead confirming the presence of ESBL genes by PCR. Phenotypic detection methods alone may identify intrinsically 3GCR isolates or those that falsely suggest ESBL production [24–26]. AmpC genes were identified in 15% isolates assessed (17.5% specimens), with most (22/23) being inherently chromosomal in location [26]. Notably, however, the sole E. coli isolate identified contained a plasmid-mediated CMY-2 which was potentially transferable.
The fact that resistance to anti-folate agents was the most common resistance phenotype identified among potential ESBL-producing strains and was noted in 68.3% of all CD/PR specimens is important, given that trimethoprim-sulfamethoxazole is widely used in pork production and some chicken farms [15, 16]. Hence these results may be no surprise, but at least serve as a potential “wake up call” to farmers who are concerned about the consequences of frequent antibiotic use. Importantly, 9 isolates (7.5% CD/PR specimens) displayed an MDR phenotype, with only one strain (Bordetella trematum) being resistant to fluoroquinolones – consistent with Australia’s strict controls on fluoroquinolone use in agriculture and similar to previous studies on this issue [14].
Acinetobacter, Serratia, Hafnia and Pseudomonas spp. are all known to be common in the environment and to be present on some fruit and vegetables [36, 37], but their presence may be a potential source of resistance genes [38].
Given the uncertainty about which testing regimen would be ideally suited for a large national food safety screening program for MDR contamination [9–13], we believe our methodology was a practical approach that is potentially relevant and meaningful to retail consumers and which could be up-scaled without the need for major infrastructure or specialised training. In comparison, all previous published Australian studies have assessed non-meat items such as animal faeces or eggs [39–42].
Our findings differ from those by other authors. Overdevest et al. [8] reported that 79.8% of retail chicken meat samples in the Netherlands had organisms with ESBL genes present, while only 1.8% of pork samples grew an ESBL-producing organism. However, this study focused particularly on E. coli and K. pneumoniae without commenting on other organisms isolated. Stewardson et al. [7] reported 86% contamination of chicken meat products delivered to a tertiary hospital in Switzerland with ESBL-producing Enterobacteriaceae species. Similar to our results, MDR strains were uncommon.
This study has some limitations. Firstly, the sample size of 120 specimens, while consistent with similar studies, is relatively small in the context of overall Australian supply [2, 7, 8, 43–47]. Secondly, we were not able to track the original farm source of the CD and PR products, although one might expect larger supermarket chains to have a limited number of defined contracted suppliers. Further research to investigate the rates of contamination at each step of the meat production process, including samples from animals in farms, carcasses and meat products in slaughterhouses and of meat products distributed to third party organisations for packaging and distribution, may be helpful to identify if there is a common source of contamination. Thirdly, our sample preparation (including initial 24 h culture in non-selective media), the subsequent selective culturing techniques provided enhanced sensitivity for 3GCR-GNBs but did not allow us to accurately quantify the burden of contamination in each CD/PR sample. Importantly, we did not assess for phenotypic colistin resistance since laboratory methods are evolving [48, 49], nor did we assess for mcr genes since this resistance mechanism was only first reported in 2016 [50]. Notably, colistin resistance appears to be currently rare in Australia [51, 52] and colistin is infrequently used in Australian agriculture [2, 53]. Finally, Australia does not import fresh chicken meat, nor any fresh bone-containing pork products [13, 17], which means that all of our specimens came from animals born and grown in Australia. As such we cannot comment on any possible difference in contamination between these Australian products and similar, but boned, imported chicken and pork processed meat products.
We believe our findings raise important questions regarding future food testing programs and potentially highlight the importance of routine public health measures related to safe food preparation such as appropriate hand hygiene before/after handling uncooked meat products, adequate washing of kitchen utensils and surfaces that have contact with uncooked meat and appropriate cooking methods to ensure destruction of any contaminating bacteria. These public health messages may be of particular importance to patient groups where immunosuppression is likely, such as those with haematological malignancy or transplant recipients. Further research into the potential source(s) of retail meat contamination is warranted.
Conclusion
Overall, we found low rates of MDR-GNBs in Australian chicken and pork meat, but potential 3GCR-GNBs are common (93% specimens), as is resistance to trimethoprim-sulfamethoxazole. Food testing programs that only assess for E. coli are likely to severely underestimate the diversity of 3GCR organisms in fresh meat.
References
Davies S. (2013). Annual report of the chief medical officer, 2011 – volume 2. https://www.gov.uk/government/publications/chief-medical-officer-annual-report-volume-2 Accessed 16th July 2016.
Australian Government. Department of Health, Department of Agriculture (2015). Responding to the Threat of Antimicrobial Resistance. https://www.health.gov.au/internet/main/publishing.nsf/.../amr-strategy-2015-2019.pdf. Accessed 8th Nov 2016.
O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. London: H M Government/Wellcome Trust; 2016. p. 2016.
WHO – World Health Organization (2012).The evolving threat of antimicrobial resistance – Options for action. http://www.who.int/patientsafety/implementation/amr/publication/en/ Accessed 8th Nov 2016.
WHO (2015). Global action plan on antimicrobial resistance. Geneva: World Health Organization, 2015. http://www.who.int/drugresistance/global_action_plan/en/ Accessed 8th Nov 2016.
Kluytmans JA, Overdevest IT, Willemsen I, et al. Extended-spectrum β-lactamase-producing Escherichia Coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis. 2013;56(4):478–87.
Stewardson AJ, Renzi G, Maury N, et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in hospital food: a risk assessment. Infect Control Hosp Epidemiol. 2014;35(4):375–83.
Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. Extended-Spectrum ß-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis. 2011;17(7):1216–22.
EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) (2015). EU Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA Journal 2015;13:4036. http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-zoonotic-bacteria-humans-animals-food-EU-summary-report-2013.pdf. Accessed 8th Nov 2016.
DANMAP (2014). DANMAP 2013 - use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. September 2015. Copenhagen. http://www.danmap.org/~/media/Projekt%20sites/Danmap/DANMAP%20reports/DANMAP%202014/Danmap_2014.ashx. Accessed 8th Nov 2016.
ECDC (European Centre for Disease Prevention and Control), EFSA (European Food Safety Authority) and EMA (European Medicines Agency) (2015). ECDC/EFSA/EMA first joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. Stockholm/Parma/London: ECDC/EFSA/EMA, 2015. EFSA Journal 13: 4006. https://www.efsa.europa.eu/en/efsajournal/pub/4006 Accessed 8th Nov 2016.
Guerra B, Fischer J, Helmuth R. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol. 2014;171:290.
JETACAR (Joint Expert Advisory Committee on Antibiotic Resistance [JETACAR]). The use of antibiotic in food producing animals: antibiotic-resistant bacteria in animals and humans. 1999. Commonwealth of Australia. http://www.health.gov.au/internet/main/publishing.nsf/Content/health-pubs-jetacar-cnt.htm/$FILE/jetacar.pdf. Accessed 6th Mar 2015.
Cheng AC, Turnidge J, Collignon P, Looke D, Barton M, Gottlieb T. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg Infect Dis. 2012;18(9):1453–60.
APVMA - Australian Pesticides and Veterinary Medicines Authority (2014). Quantity of antimicrobial products sold for veterinary use in Australia. Australian Pesticides and Veterinary Medicines Authority. Kingston. Australia. https://apvma.gov.au/node/11816. Accessed 8th Nov 2016.
Jordan D, Chin JJ, Fahy VA, Barton MD, Smith MG, Trott DJ. Antimicrobial use in the Australian pig industry: results of a national survey. Aust Vet J. 2009;87(6):222–9.
Australia - Pork meat import restrictions (PRRS/PMWS). European Commission Trade, Market Access Database. Agriculture and Fisheries Sector. 2012 May 2.
Government of Canada. Sample Collection, Preparation & Laboratory Methodologies. January 2010. National Integrated Enteric Disease Surveillance Program. http://www.phac-aspc.gc.ca/foodnetcanada/niedsp10-pnisme10/s02-eng.php Accessed 8th Nov 2016.
Kanki M, Sakata J, Taguchi M, Kumeda Y, Ishibashi M, Kawai T, Kawatsu K, Yamasaki W, Inoue K, Miyahara M. Effect of sample preparation and bacterial concentration on Salmonella enterica detection in poultry meat using culture methods and PCR assaying of preenrichment broths. Food Microbiol. 2009;26:1–3.
Clinical and Laboratory Standards Institute (CLSI). M100-S25 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fifth informational supplement. January 2015. Vol 35, No. 3.
European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST Expert Rules Version 3.1. Intrinsic Resistance and Exceptional Phenotypes Tables. http://www.eucast.org/expert_rules_and_intrinsic_resistance/ (Accessed 30th Jan 2018).
European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrobial wild type distributions of microorganisms. https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=-1&Specium=430. (Accessed 29th Jan 2018).
European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrobial wild type distributions of microorganisms. https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=-1&Specium=218. (Accessed 29th Jan 2018).
Jiang X, Zhang Z, Li M, et al. Detection of extended-Spectrum -lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:2990–5.
Beceiro A, Fernández-Cuenca F, Ribera A, et al. False extended-spectrum beta-lactamase detection in Acinetobacter spp. due to intrinsic susceptibility to clavulanic acid. J Antimicrob Chemother. 2008;61:301–8.
Thomson KS. Extended-Spectrum-β-lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol. 2010;48:1019–25.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Hanson ND, Thomson KS, Moland ES, Sanders CC, Berthold G, Penn RG. Molecular characterization of a multiply resistant Klebsiella pneumoniae encoding ESBLs and a plasmid-mediated AmpC. J Antimicrob Chemother. 1999;44:377–80.
Chia JH, Chu C, Su LH, Chiu CH, Kuo AJ, Sun CF, et al. Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M β-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J Clin Microbiol. 2005;43:4486–91.
Birkett CI, Ludlam HA, Woodford N, Brown DFJ, Brown NM, Roberts MTM, et al. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum β-lactamases. J Med Microbiol. 2007;56(Pt 1):52–5.
Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62.
Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22.
Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007;45:544–7.
Clausen PT, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J Antimicrob Chemother. 2016;71(9):2484–8.
Calbo E, Freixas N, Xercavins M, et al. Foodborne nosocomial outbreak of SHV1 and CTX-M-15-producing Klebsiella Pneumoniae: epidemiology and control. Clin Infect Dis. 2011;52(6):743–9.
Doughari HJ, Ndakidemi PA, Human IS, Benade S. The ecology, biology and pathogenesis of Acinetobacter spp.:an overview. Microbes Environ. 2011;26:101–12.
Janda JM, Abbott SL. The genus hafnia: from soup to nuts. Clin Microbiol Rev. 2006;19:12–8.
Argudín MA, Deplano A, Meghraoui A, et al. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics (Basel). 2017;6(2):E12.
van Breda LK, Dhungyel OP, Ward MP. Antibiotic resistant Escherichia coli in southeastern Australian pig herds and implications for surveillance. Zoonoses Public Health. 2018;65(1):e1–7.
Obeng AS, Rickard H, Ndi O, Sexton M, Barton M. Antibiotic resistance, phylogenetic grouping and virulence potential of Escherichia Coli isolated from the faeces of intensively farmed and free range poultry. Vet Microbiol. 2012;54(3–4):305–15.
Pande VV, Gole VC, McWhorter AR, Abraham S, Chousalkar KK. Antimicrobial resistance of non-typhoidal salmonella isolates from egg layer flocks and egg shells. Int J Food Microbiol. 2015;203:23–6.
Smith MG, Jordan D, Gibson JS, Cobbold RN, Chapman TA, Abraham S, Trott DJ. Phenotypic and genotypic profiling of antimicrobial resistance in enteric Escherichia Coli communities isolated from finisher pigs in Australia. Aust Vet J. 2016;94(10):371–6.
Folster JP, Pecic G, Singh A, Dvual B, Rickert R, Ayers S, Abbott J, McGlinchey B, Bauer-Turpin J, Haro J, Hise K, Zhao S, Fedorka-Cray PJ, Whichard J, McDermott PF. Characterization of extended-Spectrum cephalosporin-resistant Salmonella enterica Serovar Heidelberg isolated from food animals, retail meat, and humans in the United States 2009. Foodborne Pathog Dis. 2012;9(7):638–45.
Mihaiu L, Lapusan A, Tanasuica R, Sobolu R, Mihaiu R, Oniga O, Mihaiu M. First study of Salmonella in meat in Romania. J Infect Dev Ctries. 2014;8(1):50–8.
Ojer-Usoz E, Gonzalez D, Vitas AI, et al. Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013;93(2):316–21.
Silva N, Costa L, Goncalves A, Sousa M, Radhouani H, Brito F, et al. Genetic characterisation of extended-spectrum beta-lactamases in Escherichia Coli isolated from retail chicken products including CTX-M-9 containing isolates: a food safety risk factor. Br Poult Sci. 2012;53(6):747–55.
Egea P, Lopez-Cerero L, Navarro MD, Rodriguez-Bano J, Pascual A. Assessment of the presence of extended-spectrum beta-lactamase-producing Escherichia Coli in eggshells and ready-to-eat products. Eur J Clin Microbiol Infect Dis. 2011;30(9):1045–7.
Matuschek E, Åhman J, Webster C, Kahlmeter G. Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect. 2017;17:30667–5.
Carretto E, Brovarone F, Russello G, et al. Clinical validation of the SensiTest™ Colistin, a broth microdilution based method to evaluate colistin MICs. J Clin Microbiol. 2018:01523–17. https://doi.org/10.1128/JCM.01523-17. [Epub ahead of print]
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8.
Hadjadj L, Riziki T, Zhu Y, Li J, Diene SM, Rolain JM. Study of mcr-1 gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes (Basel). 2017;8(12):E394.
Ellem JA, Ginn AN, Chen SC, et al. Locally Acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013. Emerg Infect Dis. 2017;23(7):1160–3.
Australian Commission on Safety and Quality in Health Care. AURA 2016: first Australian report on antimicrobial use and resistance in human health. Commonwealth of Australia, 2016. https://www.safetyandquality.gov.au/publications/aura-2016-first-australian-report-on-antimicroibal-use-and-resistance-in-human-health/ (Accessed 29th Jan 2018).
Acknowledgements
We are grateful to John Bourke for his assistance with education regarding the Australian pork industry and Ms. Trudi Bannam for technical laboratory assistance.
Funding
No specific funding was received for this project – it was supported internally by the Infectious Diseases Department, Austin Health.
Author information
Authors and Affiliations
Contributions
JEM, AJP, MB and MLG designed the study, including approach to specimen sampling, handling and laboratory methods used. JEM, AJP, SAB, EAG and JMB undertook the laboratory testing. JEM, AJP and MLG undertook any necessary statistical analyses and prepared the manuscript with feedback/input from all other authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Figure S1. Location of 30 meat retailers in relation to Austin Hospital in eastern Melbourne. (DOCX 2275 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
McLellan, J.E., Pitcher, A.J., Ballard, S.A. et al. Superbugs in the supermarket? Assessing the rate of contamination with third-generation cephalosporin-resistant gram-negative bacteria in fresh Australian pork and chicken. Antimicrob Resist Infect Control 7, 30 (2018). https://doi.org/10.1186/s13756-018-0322-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-018-0322-4