Abstract
Background
Many outbreaks due to Serratia marcescens among neonates have been described in the literature but little is known about the role of whole genome sequencing in outbreak analysis and management.
Methods
Between February and March 2013, 2 neonates and 2 infants previously hospitalised in the neonatal unit of a tertiary care centre in Switzerland, were found to be colonised with S. marcescens. An investigation was launched with extensive environmental sampling and neonatal screening in four consecutive point prevalence surveys between April and May 2013. All identified isolates were first investigated by fingerprinting and later by whole genome sequencing. Audits of best practices were performed and a hand hygiene promotion programme was implemented.
Results
Twenty neonates were colonised with S. marcescens. No invasive infection due to S. marcescens occurred. All 231 environmental samples were negative. Hand hygiene compliance improved from 51% in April 2013 to 79% in May 2013 and remained high thereafter. No S. marcescens was identified in point prevalence surveys in June and October 2013. All strains were identical in the fingerprinting analysis and closely related according to whole genome sequencing.
Conclusions
Improving best practices and particularly hand hygiene proved effective in terminating the outbreak. Whole genome sequencing is a helpful tool for genotyping because it allows both sufficient discrimination of strains and comparison to other outbreaks through the use of an emerging international database.
Similar content being viewed by others
Background
Serratia marcescens has long been recognized as an important pathogen in neonatal intensive care units (NICUs). It is the third most common pathogen identified in published NICU outbreaks [1], and it has been found to account for 15% of all culture-positive nosocomial infections in this setting [2]. The large number of outbreak reports underestimates the true occurrence of S. marcescens in neonatology units and NICUs. According to the results of the mandatory surveillance of healthcare-associated infections (HAIs) in very low birth weight infants in Germany from 2006 to 2011, at least one to two Serratia outbreaks per year are expected for Germany alone [3].
S. marcescens causes a wide range of clinical manifestations in neonates, from asymptomatic colonization to infections such as urinary tract infections, pneumonia, sepsis or meningitis [4, 5]. Risk factors for Serratia spp. acquisition by neonates are related to immaturity, prolonged hospital stay, antibiotic use, and mechanical ventilation [4, 6].
The objective of this outbreak report was to summarize the investigation and successful management of a S. marcescens outbreak in neonates and to investigate the contribution of using whole genome sequencing. This report follows the ORION (Outbreak Reports and Intervention Studies Of Nosocomial infection) statement [7].
Methods
Setting
The University of Geneva Hospitals (HUG), are a 1′800-bed primary and tertiary care center with about 47,000 admissions accounting for 660,000 patient-days per year. At the time of the outbreak it offered 17 places in the NICU, and 12 places in the geographically separate pediatric intensive care unit (PICU), where neonates are cared for when mechanical ventilation is needed. Neonates who are clinically stable but need prolonged stay for non-medical reasons are transferred to the unit for child development (UCD).
Outbreak
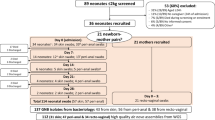
The outbreak started in February 2013 and ended in June 2013 (Fig. 1). Between February and March 2013, two neonates in the NICU and 2 infants in the UCD were found with S. marcescens (2 vascular catheters, 2 eye swabs, and 1 urine sample). Given the organizational ties and the patient flow between NICU, PICU, and UCD, an investigation was launched by a first point prevalence survey in the three units (Fig. 2) on 23 April 2013. Eight out of 41 screened children were identified as cases in this survey, one in the PICU, 3 in the UCD and 4 in the NICU. All new cases were neonates with a present or past history of stay in the NICU. Based on these findings, we focused further activity on the NICU with extensive environmental sampling and neonatal screening during 4 consecutive prevalence surveys.
Outbreak management
Audits of best practices in the use of surface disinfectants, ointments and cosmetic products were performed, as well as weekly direct hand hygiene observations (439 opportunities in total). An intensive hand hygiene promotion programme was offered to the staff working in the NICU: daily presence in the NICU, education and training, and weekly hand hygiene audits with individual feedback.
Microbiological investigation/genotyping
In 2013, the genomes of the S. marcescens isolates obtained in the prevalence surveys were tested by a commercial fingerprinting assay (DiversiLab®, Biomerieux, France). In 2016, the genomes S. marcescens isolates were fully sequenced using an Illumina HiSeq 2500 sequencer as previously described [8].
Results
A total of 232 neonatal screenings (117 stool samples, 115 nasal swabs) [9] were performed. In addition to the 4 index cases and the 8 cases identified in the first prevalence survey, an additional three, four and one cases were identified in the second, third and fourth prevalence surveys in April and May, respectively (Fig. 2). The proportions of positive rectal and nasal swabs were 11.1% (13/117) and 6.1% (7/115), respectively. Five infants had a positive result for both rectal and nasal swabs. All cases were preterm-, and seven were very-low-birth weight infants. No new S. marcescens cases were identified until October 2016, including two further prevalence surveys, which were performed in June and October. There was no invasive infection due to S. marcescens, neither during the outbreak nor until end of December 2013. A total of 231 environmental samplings were performed (Table 1). All tested products, materials and surfaces were negative for S. marcescens. Hand hygiene compliance improved from 51% in April 2013 to 79% in May 2013 following the promotion programme. Compliance remained above 65% in the following months (Fig. 1).
Fingerprinting of 24 isolates from 16 neonates showed identical strains (Fig. 3). Whole genome sequencing was carried out on the set of 24 S. marcescens strains (Sm01-Sm24) belonging to the outbreak. The sequencing on an Illumina HiSeq produced on average a total of 13 million 150 bp reads per sample, exhibiting very high theoretical coverage values (between 300 and 900 fold). Assembly was achieved using SPAdes 3.9.0 software, after read quality trimming and filtering was applied to the raw reads. The assembly produced very similar results for each strain from the outbreak, resulting in an average genome size of 5′080’124 bp with a standard deviation of 1097 bp. This shows extreme similarity between the strains of the outbreak. Moreover, pairwise comparisons using MUMmer software were performed between each couple of strains and showed similarity above 99.999% [10]. Figure 4 shows an unrooted tree displaying all the isolates from the outbreak (Sm01-24), and one representative strain isolated in Germany during a SM outbreak in 2016 (SMB2099) [11].
Discussion
This report suggests that focusing on and improving hand hygiene is effective in terminating a S. marcescens outbreak among neonates. Whole genome sequencing is a useful tool for outbreak investigation because it is more discriminative than standard fingerprinting tests and allows benchmarking with other outbreaks thanks to a growing gene bank.
The source of S. marcescens outbreaks in NICUs is rarely identified. A review summarizing 48 NICU outbreaks reported an identified source in only 40% (19/48), the most frequent being a colonized or infected index patient (8/48), followed by equipment for patient care (6/48), an (unspecified) environment (4/48), and food (1/48) [12]. Colonized or infected patients are thought to represent the most important reservoir of S. marcescens outbreaks in NICUs [4, 13]. In the currently described situation, the investigation revealed two additional NICU cases prior to the outbreak. A mother of one of them hadan amnion infection syndrome due to S. marcescens three months before the start of the outbreak, and thus, may have been the most likely source [12]. In the literature, healthcare workers were never identified as a source, but few studies investigated this specifically [14]. Like in our study, extensive environmental screening rarely yielded positive results in the reported outbreaks, suggesting that environmental contamination may play a limited role.
The clinical implication of colonization with S. marcescens in neonates and its implication in outbreaks is not always clear. A number of studies reported that S. marcescens is rather commonly isolated in stool of preterm neonates within the first weeks of life [6, 15, 16]. However, the pathogen was not isolated consistently in preterm neonates either [17], and S. marcescens has not been isolated in healthy term babies [18]. Thus, while the finding of S. marcescens in neonatal stool can be considered a regular event in NICUs, the pathogen per se is not part of the normal early gut flora of healthy newborns in the community. Once the intestines of a neonate are colonized with S. marcescens from the (pathologic) outside, they can become the “source” of an outbreak. This makes control of Serratia outbreaks very difficult, and underlines the importance of hands in the transmission of the pathogen and the possible successful management of an outbreak.
Already in 1981, contaminated hand-washing brushes were reported to be implicated in a S. marcescens outbreak, indirectly pointing to the pivotal role of hand hygiene in the spread of the pathogen [19]. Contaminated soaps were identified in another report [20]. Interventions to control outbreaks included always both hand hygiene and environmental control [9, 21,22,23,24,25,26,27,28]. Given that all environmental samples were negative in our outbreak investigation and work had been done in the past to dedicate equipment, drugs, ointments and other cosmetic products to individual neonates (avoiding multivials) [29], our intervention focused on hand hygiene improvement. This was all the more justified because a decrease of hand hygiene compliance had been identified as part of the investigation; hand hygiene compliance was 72% in the year before the outbreak.
Whole genome sequencing technology emerged as a powerful tool to identify clonality among outbreak isolates. The genome of a S. marcescens strain that sparked an outbreak among infants in Germany was reported very recently [11, 12]. Comparison with our index case revealed a moderate difference around 200 SNPs. Other isolates identified from our institution and not related to the outbreak were sequenced and the number of SNPs was higher than 100′000 (data not shown).
There are limitations in our outbreak investigation. First, environmental sampling was large but we did not consistently use neutralizing agents for testing the different cosmetic products, resulting in potential underreporting. However, cosmetic products were strictly confined to the neonates and no multivials were in use. A positive finding of a product with a case would not have been prove of the product being the source. Second, we did not produce formal records from the audits other than hand hygiene.
Conclusions
Improving best practice and particularly hand hygiene are effective in terminating the outbreak. This highlights the role of hand hygiene in the cause but also in the successful management of S. marcescens outbreaks in neonates. Whole genome sequencing is a helpful tool for genotyping because it allows both sufficient discrimination of strains within the outbreak and comparison to other outbreaks through an emerging international database.
Change history
16 January 2018
A Correction to this paper has been published: https://doi.org/10.1186/s13756-017-0295-8
Abbreviations
- HAI:
-
Healthcare-associated infection
- HUG:
-
University of Geneva hospitals
- NICU:
-
Neonatal intensive care unit
- ORION:
-
Outbreak reports and intervention studies of nosocomial infection
- PICU:
-
Pediatric intensive care unit
- SNP:
-
Single nucleotide polymorphism
- UCD:
-
Unit for child development
References
Gastmeier P, Loui A, Stamm-Balderjahn S, Hansen S, Zuschneid I, Sohr D, Behnke M, Obladen M, Vonberg RP, Ruden H. Outbreaks in neonatal intensive care units - they are not like others. Am J Infect Control. 2007;35(3):172–6.
Raymond J, Aujard Y. Nosocomial infections in pediatric patients: a European, multicenter prospective study. European study group. Infect Control Hosp Epidemiol. 2000;21(4):260–3.
Schwab F, Geffers C, Piening B, Haller S, Eckmanns T, Gastmeier P. How many outbreaks of nosocomial infections occur in German neonatal intensive care units annually? Infection. 2014;42(1):73–8.
Voelz A, Muller A, Gillen J, Le C, Dresbach T, Engelhart S, Exner M, Bates CJ, Simon A. Outbreaks of Serratia Marcescens in neonatal and pediatric intensive care units: clinical aspects, risk factors and management. Int J Hyg Environ Health. 2010;213(2):79–87.
David MD, Weller TM, Lambert P, Fraise AP. An outbreak of Serratia Marcescens on the neonatal unit: a tale of two clones. J Hosp Infect. 2006;63(1):27–33.
Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, Fernandez L, Rodriguez JM, Jimenez E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One. 2013;8(6):e66986.
Stone SP, Cooper BS, Kibbler CC, Cookson BD, Roberts JA, Medley GF, Duckworth G, Lai R, Ebrahim S, Brown EM, et al. The ORION statement: guidelines for transparent reporting of outbreak reports and intervention studies of nosocomial infection. Lancet Infect Dis. 2007;7(4):282–8.
Von Dach E, Diene SM, Fankhauser C, Schrenzel J, Harbarth S, Francois P. Comparative genomics of community-associated Methicillin-resistant Staphylococcus Aureus shows the emergence of clone ST8-USA300 in Geneva, Switzerland. J Infect Dis. 2016;213(9):1370–9.
Steppberger K, Walter S, Claros MC, Spencker FB, Kiess W, Rodloff AC, Vogtmann C. Nosocomial neonatal outbreak of Serratia Marcescens--analysis of pathogens by pulsed field gel electrophoresis and polymerase chain reaction. Infection. 2002;30(5):277–81.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12.
Serratia marcescens SMB2099 complete genome; GenBank: HG738868.1. https://www.ncbi.nlm.nih.gov/nuccore/HG738868.1. Accessed 1 Dec 2017.
Gastmeier P. Serratia Marcescens: an outbreak experience. Front Microbiol. 2014;5:81.
Duggan TG, Leng RA, Hancock BM, Cursons RT. Serratia Marcescens in a newborn unit--microbiological features. Pathology. 1984;16(2):189–91.
Assadian O, Berger A, Aspock C, Mustafa S, Kohlhauser C, Hirschl AM. Nosocomial outbreak of Serratia Marcescens in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2002;23(8):457–61.
Moles L, Gomez M, Jimenez E, Fernandez L, Bustos G, Chaves F, Canton R, Rodriguez JM, Del Campo R. Preterm infant gut colonization in the neonatal ICU and complete restoration 2 years later. Clin Microbiol Infect. 2015;21(10):e931–10. 936
Drell T, Lutsar I, Stsepetova J, Parm U, Metsvaht T, Ilmoja ML, Simm J, Sepp E. The development of gut microbiota in critically ill extremely low birth weight infants assessed with 16S rRNA gene based sequencing. Gut Microbes. 2014;5(3):304–12.
Aujoulat F, Roudiere L, Picaud JC, Jacquot A, Filleron A, Neveu D, Baum TP, Marchandin H, Jumas-Bilak E. Temporal dynamics of the very premature infant gut dominant microbiota. BMC Microbiol. 2014;14:325.
Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177.
Anagnostakis D, Fitsialos J, Koutsia C, Messaritakis J, Matsaniotis N. A nursery outbreak of Serratia Marcescens infection. Evidence of a single source of contamination. Am J Dis Child. 1981;135(5):413–4.
Archibald LK, Corl A, Shah B, Schulte M, Arduino MJ, Aguero S, Fisher DJ, Stechenberg BW, Banerjee SN, Jarvis WR. Serratia Marcescens outbreak associated with extrinsic contamination of 1% chlorxylenol soap. Infect Control Hosp Epidemiol. 1997;18(10):704–9.
Al Jarousha AM, El Qouqa IA, El Jadba AH, Al Afifi AS. An outbreak of Serratia Marcescens septicaemia in neonatal intensive care unit in Gaza City Palestine. J Hosp Infect. 2008;70(2):119–26.
Bates CJ, Pearse R. Use of hydrogen peroxide vapour for environmental control during a Serratia outbreak in a neonatal intensive care unit. J Hosp Infect. 2005;61(4):364–6.
Braver DJ, Hauser GJ, Berns L, Siegman-Igra Y, Muhlbauer B. Control of a Serratia Marcescens outbreak in a maternity hospital. J Hosp Infect. 1987;10(2):129–37.
Buffet-Bataillon S, Rabier V, Betremieux P, Beuchee A, Bauer M, Pladys P, Le Gall E, Cormier M, Jolivet-Gougeon A. Outbreak of Serratia Marcescens in a neonatal intensive care unit: contaminated unmedicated liquid soap and risk factors. J Hosp Infect. 2009;72(1):17–22.
Casolari C, Pecorari M, Fabio G, Cattani S, Venturelli C, Piccinini L, Tamassia MG, Gennari W, Sabbatini AM, Leporati G, et al. A simultaneous outbreak of Serratia Marcescens and Klebsiella Pneumoniae in a neonatal intensive care unit. J Hosp Infect. 2005;61(4):312–20.
Gillespie EE, Bradford J, Brett J, Kotsanas D. Serratia Marcescens bacteremia - an indicator for outbreak management and heightened surveillance. J Perinat Med. 2007;35(3):227–31.
Jang TN, Fung CP, Yang TL, Shen SH, Huang CS, Lee SH. Use of pulsed-field gel electrophoresis to investigate an outbreak of Serratia Marcescens infection in a neonatal intensive care unit. J Hosp Infect. 2001;48(1):13–9.
Sarvikivi E, Lyytikainen O, Salmenlinna S, Vuopio-Varkila J, Luukkainen P, Tarkka E, Saxen H. Clustering of Serratia Marcescens infections in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2004;25(9):723–9.
Pessoa-Silva CL, Hugonnet S, Pfister R, Touveneau S, Dharan S, Posfay-Barbe K, Pittet D. Reduction of health care associated infection risk in neonates by successful hand hygiene promotion. Pediatrics. 2007;120(2):e382–90.
Acknowledgments
We would like to thank Delphine Scalia-Perreard, Claude Ginet, and Sylvie Touveneau for the outbreak investigation.
Funding
No external funding.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
BH, IS, and WZ organised and performed the investigation and management of the outbreak. DB, GR and PF performed the microbiological investigation and whole genome sequencing. WZ, DB, and PF did the data analysis. WZ wrote the first draft of the manuscript. WZ, IS, DB, BH, RP, GR, DP, JS, and PF reviewed and contributed to subsequent drafts. All authors approved the final version for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable (outbreak report; no interventions humans).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional information
The original article has been revised. The title was corrected to remove an errant EM ID.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zingg, W., Soulake, I., Baud, D. et al. Management and investigation of a Serratia marcescens outbreak in a neonatal unit in Switzerland – the role of hand hygiene and whole genome sequencing. Antimicrob Resist Infect Control 6, 125 (2017). https://doi.org/10.1186/s13756-017-0285-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-017-0285-x