Abstract
Background
We report on an outbreak in a surgical, interdisciplinary intensive care unit (ICU) of a tertiary care hospital. We detected a cluster of ICU patients colonized or infected with multidrug-resistant Pseudomonas aeruginosa. We established an outbreak investigation team, performed an exploratory epidemiological analysis and initiated an epidemiology-based intervention.
Methods
As part of the outbreak investigation, we performed microbiological examinations of the sinks in the patient rooms and a retrospective case-control study. All patients admitted to the outbreak ICU between January 2012 and February 2014 were included. Cases were patients colonized with the outbreak strain. Controls were patients with a different Pseudomonas aeruginosa strain. Risk factors were evaluated using multivariable conditional logistic regression analysis. Strain typing was performed using the repetitive element-based polymerase chain reaction (rep-PCR) DiversiLab system.
Results
The outbreak strain was found in the sinks of five (of 16) patient rooms. Altogether 21 cases and 21 (randomly selected) controls were included. In the univariate analysis, there was no significant difference in baseline data of the patients. In the multivariate analysis, stay in a room with a colonized sink (Odds Ratio[OR] 11.2, p = 0.007) and hemofiltration (OR 21.9, p = 0.020) were independently associated with an elevated risk for colonization or infection by the outbreak strain. In a subsequent evaluation of the work procedures associated with hemofiltration, we found that the ultra-filtrate bags had been on average five times per day emptied in the sinks of the patient rooms and were used multiple for the same patient. We exchanged the traps of the contaminated sinks and eliminated work procedures involving sinks in patient rooms by implementation of single use bags, which are emptied outside patient rooms to reduce splash water at the sinks. In the 20 month follow-up period, the outbreak strain was detected only once, which indicated that the outbreak had been ceased (incidence 0.75% vs. 0.04%, p < 0.001) Furthermore, the incidence of Pseudonomas aeruginosa overall was significantly decreased (2.5% vs. 1.5%, p < 0.001).
Conclusion
In ICUs, limiting work processes involving sinks results in reduced multidrug-resistant Pseudomonas aeruginosa rates. ICUs with high rates of Pseudomonas aeruginosa should consider eliminating work processes that involve sinks and potentially splash water in close proximity to patients.
Trial registration
All data were surveillance based data which were obtained within the German Law on Protection against Infection (“Infektionsschutzgesetz”). Therefore a trial registration was not required.
Similar content being viewed by others
Background
Multidrug-resistant (MDR) P. aeruginosa are among the most commonly-found organisms which cause nosocomial infections in intensive care units [12, 13, 18, 21]. They are associated with increased mortality as well as increased hospital costs [14]. P. aeruginosa is a hydrophilic Gram-negative rod often found in water drainage systems in hospitals [17]. The organism has further been described as extremely adaptable to selective pressure caused by antimicrobial agents [13]. Due to the substantial necessity of antimicrobial therapy in intensive care units (ICU), the selective pressure in this setting is considerably high. This is supported by the finding that antimicrobial therapy prior to an ICU stay increases the risk for colonization with MDR P. aeruginosa [8, 15]. Once the organisms are introduced into an ICU, they can cause outbreaks that are often associated with sinks or faucets as a continuous source of further spread [1, 2, 11, 19, 20]. However, the most likely transmission route is direct person-to-person contact [10].
We report on an outbreak of MDR P. aeruginosa in an interdisciplinary ICU of a tertiary care, university hospital. The unit was comprised of 16 rooms with 30 intensive care beds. Within the scope of our infection control assignment, we collected all multidrug resistant organisms (MDRO) isolated from clinical or screening cultures and stored them in our MDRO bank. Whenever an epidemiological link between these MDRO isolates is suspected, molecular strain typing is performed. By this means, a cluster of MDR P. aeruginosa strains was observed in December 2013. Because the strain typing results suggested a clonal relationship, we subsequently re-cultured earlier MDR P. aeruginosa isolates from the frozen MDRO bank of our institute since January 2012. Furthermore, we decided to initiate an investigation to control the outbreak, find the transmission route, eliminate the potential source, and avoid future clusters.
Methods
The outbreak and the subsequent analysis was performed in a tertiary care university hospital with over 3000 beds. The outbreak ward was a surgical intensive care unit with 30 beds. All beds were equipped for intubation and ventilator support. The patient beds were distributed over 16 rooms. Each room was equipped with one clinical hand hygiene sink (Fig. 1). The sinks consisted of ceramic washbasin with chrome-plated tap made of metal and an attached downstream bacterial filter. Together with the rest of the room, the surfaces of the sinks were cleaned at least twice daily. The Sinks were also used for bathing and grooming the patients.
For the suspected outbreak, an outbreak investigation team was established comprised of two infection control physicians, one infection control nurse, the attending intensive care physician of the ward, and a microbiologist. Because all MDRO isolates were routinely collected and frozen, we were able to examine retrospectively all MDR P. aeruginosa isolates from the affected ward for the 2 years prior to the start of the outbreak investigation. P. aeruginosa isolates detected at least 72 h after admission to the ward were classified as ward-associated.
Microbiological methods and clinical and environmental sampling
Clinical and environmental samples including identification on species level and antimicrobial susceptibility testing were performed by the VITEK 2 system (bioMérieux) and were interpreted according to European Committee on Antimicrobial Susceptibility Testing definitions (EUCAST, http://www.eucast.org). Phenotypical categorization of the outbreak strain as MDR (multidrug-resistant) was performed using standard definitions [13]. Rep-PCR was performed on all available clinical and environmental MDR P. aeruginosa isolates derived from the ICU between 1st January 2012 and December 31, 2015. Isolates with rep-PCR profiles yielding a similarity of >95% were considered clonally related [6]. In order to detect outbreak clusters, we performed rep-PCR of all MDR P. aeruginosa isolates from clinical specimens of the respective ward between 1st January 2012 and the intervention on April 30, 2014. As a follow up, the analysis was repeated until December 31, 2015 comparing new MDR P. aeruginosa strains to a reference outbreak strain from the initially discovered cluster. In order to assess the likelihood of environmental contamination and subsequent spread, the sinks of all patient rooms in the respective ward were probed using sterile cotton swabs. The MDR P. aeruginosa isolates collected environmentally were also compared to the reference outbreak rep-PCR profile. Rep-PCR was performed with the DiversiLab-System (BioMerieux) [5], using Pearson correlation coefficient pairwise pattern matching and the unweighted pair group method with arithmetic mean (UPGMA) clustering algorithm.
Exploratory case-control-study
We conducted a retrospective case-control-study. The initial base cohort was comprised of all patients that were found (for the first time during their hospital stay) to be colonized or infected with P. aeruginosa 48 h after admission to the ICU. As a result these strains were considered ward-associated. All patients admitted to the outbreak ICU during the ongoing outbreak between 1st January 2012 and 30th April 2014 were included. Cases were defined as patients with either a colonization or infection by the MDR P. aeruginosa outbreak strain. Controls were patients colonized or infected with any P. aeruginosa strain other than the outbreak strain. The controls were randomly selected from the respective sub-cohort of all potential controls (n = 78). Risk factors for colonization were collected by means of a retrospective analysis of the patients’ records. Risk factors were extracted for the period between admission to the ward and P. aeruginosa detection. The following basic epidemiological parameters were collected: age, gender, overall length of stay on the ward, stay in a single or shared room as well as following risk factors for acquisition of the outbreak strain: surgical drainage from any body site, dialysis or hemofiltration, invasive intubation, central venous catheter (CVC), time (days) before P. aeruginosa detection, severity of diseases (SAPS II), stay in a room with a colonized sink, clinically diagnosed P. aeruginosa infection, tracheal cannula, leukopenia, administration of an immunosuppressive drug, shock as defined by intensive care medicine definitions [3], antimicrobial therapy, and contact with a patient colonized with the outbreak strain.
Statistical methods
Differences were tested by Chi-square or Wilcoxon rank sum test in a univariate analysis. A multivariable analysis was performed to estimate the effect of factors independently associated with colonization by the outbreak strain using a stepwise forward conditional logistic regression. Included in the multivariable analysis were all parameters with a p-value ≤ 0.100 in the univariate analysis. The p-values for including a variable in the multivariable model was ≤0.05 and for excluding >0.05. Odds ratios (OR) and their 95% confidence intervals (CI 95%) were calculated. All analyses were performed using SPSS (IBM).
Results
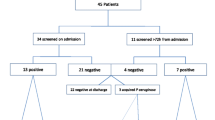
Between 1st January, 2012 and 30th April 2014 altogether 135 P. aeruginosa-colonized or infected patients were found on the respective ward. The mean length of stay in the outbreak ICU during this time period was 6.34 days. Based on the results of the rep-PCR examination, we found a cluster of 26 patients overall, colonized with the same MDR PAE strain (Fig. 2). This strain showed a phenotypical resistance to acylureidopenicillins, 3rd generation cephalosporins, quinolones and carbapenems. One MDR PAE isolate from March 2014 could not be retrieved for rep-PCR and was excluded from the analysis. The cases were more or less homogeneously scattered over the total study period of 28 months, with a period of 5 months between February and June 2012 during which no MDR PAE were observed (Fig. 2). The typing results and the epidemiological curve suggested a continuous outbreak with a single MDR PAE strain that had lasted since at least January 2012. Hence, we performed environmental examinations of all sinks located in patients’ rooms. Altogether, five sinks were colonized with MDR PAE. Rep-PCR showed that all five isolates were identical to the outbreak strain (Fig. 1).
For five of the 26 cases, sufficient data was not available at the time of the analysis. For the remaining 21 cases (81%), we conducted a case control study using one control per case. Table 1 shows the results of the univariate analysis of all collected parameters for cases and controls. There was no statistically significant difference in age, sex or severity of disease (SAPS II). Among controls, 42.9% (9/21) were colonized with a MDR P. aeruginosa different from the outbreak strain. Furthermore, 66.7% (n = 14) of the cases and 28.6% (n = 6) of the controls had been located in one of the rooms with a sink colonized with the outbreak strain (p = 0.062; see Table 1). In the multivariable analysis, stay in a room with a colonized sink and dialysis/hemofiltration showed an increased risk for acquisition of the outbreak strain (Table 2).
In order to prevent further spread of the outbreak strain, we replaced the traps of the affected sinks and implemented single use hemofiltration bags and emptying was performed outside patient rooms. Furthermore, we performed additional training in hand hygiene and education of potential transmission routes for the ward’s staff. As a follow up, we analyzed the detection rate and the incidence density of MDR P. aeruginosa in clinical specimens before and after the intervention (replacement of colonized sinks/single use bags) (Table 3). After the intervention, a significant reduction was seen in the incidence density as well as the detection rate for the outbreak strain. A decreased detection rate in overall MDR P. aeruginosa clinical specimens was observed after the intervention but also an increased incidence density of clinical specimens.
Discussion
Patient colonization, or infection with MDR P. aeruginosa in intensive care units is a common phenomenon [12]. It is important to minimize the likelihood of transmission to or between ICU patients. Intensive care units are high-risk areas for nosocomial infections. Hence, ICUs require particular infection prevention measures (different from standard care wards) in order to take into account the elevated risk of infection and transmission associated with ICU-specific working procedures (e.g. ventilator support, central-line catheters). Within the scope of an outbreak investigation due to MDR PAE in an ICU, we found evidence of a transmission route associated with working procedures at sink, in particular the use of the sinks for grooming the patients. After reducing the procedures associated with sinks in patient rooms by using single use octenidine wash cloths [7] and the instruction to empty ultra-filtrate bags of the hemofiltration in a sink outside the patient room. After the implementation the outbreak strain was detected less frequently. The increase of incidence density for all MDR PAE must be seen as a result of a post outbreak increase of microbiological screening activities. This is underlined by the significant decline in the detection rate per clinical specimen and the significantly more frequent microbiological examinations after the intervention.
Contaminated sinks or faucets have been described as sources for P. aeruginosa outbreaks in the past [1, 2, 4, 11, 19, 20]. In many outbreaks, contaminated washbasins served as continuous sources. From there, P. aeruginosa is often spread further from person to person through direct contact. Our data supports this finding. Two thirds of our cases had a stay within a room with a colonized sink. Furthermore, one third of the patients had renal replacement therapy, such as dialysis or hemofiltration. In line with national recommendations, the suggested procedure in our hospital is to dispose ultra-filtrate bags via waste rooms. While investigating on site, we found that the ultra-filtrate bags from the dialysis or hemofiltration machines where used multiple times and emptied in the nearest sink (Fig. 3). The ultra-filtrate bags had a filling volume of 10 l and were changed five times per 24 h on average. The strong statistical association of renal replacement therapy with the outbreak strain could therefore reveal an as yet not described transmission route. However, two-thirds of the case patients did not receive renal replacement therapy. Therefore, other transmission routes, such as direct person-to-person contact, were most likely also involved. The improvement of hand hygiene through focused training was a key element of our outbreak control. Also splash of water from the sink into hands of staff and contamination of medical must be taken into account. As an outbreak control measure, we changed all contaminated traps and recommended to switch to single-use ultra-filtrate bags to limit the procedures associated with sinks located in patient rooms. However, within the subsequent 20 months after the reduction of working processes associated with sinks in patient rooms, the clinical detection of the outbreak isolate as well as the incidence density of MDR P. aeruginosa declined significantly. A study by Hopmann et al. in the Netherlands showed a reduction of the MDRO rate in an ICU after removing sinks from patient rooms [9].
In the univariate analysis, prolonged stay in a shared room was associated with the colonization by the outbreak strain. This did not turn out to be an independent factor in the multivariable analysis. However, currently there is an ongoing discussion whether single rooms in ICUs have the potential to reduce nosocomial infections [16]. As only few of the patients analyzed were infected with P. aeruginosa, our contribution to this discussion is limited.
Conclusion
Sinks in patient rooms in ICUs can be potential drivers of MDR P. aeruginosa outbreaks. We were able to show that reducing work processes involving sinks in patient rooms result in reduced MDR P. aeruginosa rates. ICUs with high rates of P. aeruginosa should consider eliminating work processes involving sinks in proximity to patients.
Abbreviations
- CI:
-
Confidence intervals
- CVC:
-
Central venous catheter
- ICU:
-
Intensive care unit
- MDR PAE:
-
Multidrug-resistant Pseudomonas aeruginosa
- MDR:
-
Multidrug-resistant
- MDRO:
-
Multidrug resistant organisms
- OR:
-
Odds ration
- P. aeruginosa:
-
Pseudomonas aeruginosa
- rep-PCR:
-
Repetitive element-based polymerase chain reaction
References
Bedard E, Laferriere C, Charron D, Lalancette C, Renaud C, Desmarais N, Deziel E, Prevost M. Post-outbreak investigation of Pseudomonas aeruginosa faucet contamination by quantitative polymerase chain reaction and environmental factors affecting positivity. Infect Control Hosp Epidemiol. 2015;36:1337–43.
Blanc DS, Nahimana I, Petignat C, Wenger A, Bille J, Francioli P. Faucets as a reservoir of endemic Pseudomonas aeruginosa colonization/infections in intensive care units. Intensive Care Med. 2004;30:1964–8.
Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55.
Davis RJ, Jensen SO, Van Hal S, Espedido B, Gordon A, Farhat R, Chan R. Whole genome sequencing in real-time investigation and management of a Pseudomonas aeruginosa outbreak on a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2015;36:1058–64.
Deplano A, Denis O, Rodriguez-Villalobos H, De Ryck R, Struelens MJ, Hallin M. Controlled performance evaluation of the DiversiLab repetitive-sequence-based genotyping system for typing multidrug-resistant health care-associated bacterial pathogens. J Clin Microbiol. 2011;49:3616–20.
Doleans-Jordheim A, Cournoyer B, Bergeron E, Croize J, Salord H, Andre J, Mazoyer MA, Renaud FN, Freney J. Reliability of Pseudomonas aeruginosa semi-automated rep-PCR genotyping in various epidemiological situations. Eur J Clin Microbiol InfectDis. 2009;28:1105–11.
Gastmeier P, Kampf KP, Behnke M, Geffers C, Schwab F. An observational study of the universal use of octenidine to decrease nosocomial bloodstream infections and MDR organisms. J Antimicrob Chemother. 2016;71:2569–76.
Gomez-Zorrilla S, Camoez M, Tubau F, Periche E, Canizares RDominguez MA, Ariza J, Pena C. Antibiotic pressure is a major risk factor for rectal colonization by multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Antimicrob Agents Chemother. 2014;58:5863–70.
Hopman J, Bos R, Voss A, Kolwijck E, Sturm P, Pickkers P, Tostmann A, Hoeven H. Reduced rate of MDROs after introducing ‘water-free patient care’on a large intensive care unit in the Netherlands. Antimicrob Resist Infect Control. 2015;4:O40.
Kanayama A, Kawahara R, Yamagishi T, Goto KKobaru Y, Takano M, Morisada K, Ukimura A, Kawanishi F, Tabuchi A, Matsui T, Oishi K. Successful control of an outbreak of GES-5 extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa in a long-term care facility in Japan. J Hosp Infect. 2016;93:35–41.
Knoester M, de Boer MG, Maarleveld JJ, Claas EC, Bernards AT, de Jonge E, van Dissel JT, Veldkamp KE. An integrated approach to control a prolonged outbreak of multidrug-resistant Pseudomonas aeruginosa in an intensive care unit. Clin Microbiol Infect. 2014;20:O207–15.
Maechler F, Pena Diaz LA, Schroder C, Geffers C, Behnke M, Gastmeier P. Prevalence of carbapenem-resistant organisms and other Gram-negative MDRO in German ICUs: first results from the national nosocomial infection surveillance system (KISS). Infection. 2015;43:163–8.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81.
Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3:32.
Suarez C, Pena C, Arch O, Dominguez MA, Tubau F, Juan C, Gavalda L, Sora M, Oliver A, Pujol M, Ariza J. A large sustained endemic outbreak of multiresistant Pseudomonas aeruginosa: a new epidemiological scenario for nosocomial acquisition. BMC Infect Dis. 2011;11:272.
Teltsch DY, Hanley J, Loo V, Goldberg P, Gursahaney A, Buckeridge DL. Infection acquisition following intensive care unit room privatization. Arch Intern Med. 2011;171:32–8.
Walker J, Moore G. Pseudomonas aeruginosa in hospital water systems: biofilms, guidelines, and practicalities. J Hosp Infect. 2015;89:324–7.
Weinstein RA, Gaynes R, Edwards JR, System NNIS. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–54.
Wendel AF, Kolbe-Busch S, Ressina S, Schulze-Robbecke R, Kindgen-Milles D, Lorenz C, Pfeffer K, MacKenzie CR. Detection and termination of an extended low-frequency hospital outbreak of GIM-1-producing Pseudomonas aeruginosa ST111 in Germany. Am J Infect Control. 2015;43:635–9.
Willmann M, Bezdan D, Zapata L, Susak H, Vogel W, Schroppel K, Liese J, Weidenmaier C, Autenrieth IB, Ossowski S, Peter S. Analysis of a long-term outbreak of XDR Pseudomonas aeruginosa: a molecular epidemiological study. J Antimicrob Chemother. 2015;70:1322–30.
Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–17.
Acknowledgments
We want to thank Gerald Brennan for proof-reading the manuscript.
Funding
The study was not funded.
Availability of data and materials
The datasets generated and analysed during the current study are not publicly available as these have not been anonymized but are available from the corresponding author on reasonable request.
Authors’ contribution
All authors have made substantial contributions to the conception of study, the study design and interpretation of the data. RS, FS, PG and MD led initial study design and development. DG and MB led the data collection. RS and FS led the analysis with input from SH and AK, who also helped to draft the manuscript. All authors approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable, because all data were surveillance based data which were obtained within the German Law on Protection against Infection (“Infektionsschutzgesetz”).
Ethics approval and consent to participate
Not applicable, because all data were surveillance based data which were obtained within the German Law on Protection against Infection (“Infektionsschutzgesetz”).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Salm, F., Deja, M., Gastmeier, P. et al. Prolonged outbreak of clonal MDR Pseudomonas aeruginosa on an intensive care unit: contaminated sinks and contamination of ultra-filtrate bags as possible route of transmission?. Antimicrob Resist Infect Control 5, 53 (2016). https://doi.org/10.1186/s13756-016-0157-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-016-0157-9






