Abstract
The motility of eukaryotic cilia and flagella is modulated in response to several extracellular stimuli. Ca2+ is the most critical intracellular factor for these changes in motility, directly acting on the axonemes and altering flagellar asymmetry. Calaxin is an opisthokont-specific neuronal calcium sensor protein first described in the sperm of the ascidian Ciona intestinalis. It binds to a heavy chain of two-headed outer arm dynein in a Ca2+-dependent manner and regulates ‘asymmetric’ wave propagation at high concentrations of Ca2+. A Ca2+-binding subunit of outer arm dynein in Chlamydomonas reinhardtii, the light chain 4 (LC4), which is a Ca2+-sensor phylogenetically different from calaxin, shows Ca2+-dependent binding to a heavy chain of three-headed outer arm dynein. However, LC4 appears to participate in ‘symmetric’ wave propagation at high concentrations of Ca2+. LC4-type dynein light chain is present in bikonts, except for some subclasses of the Excavata. Thus, flagellar asymmetry-symmetry conversion in response to Ca2+ concentration represents a ‘mirror image’ relationship between Ciona and Chlamydomonas. Phylogenetic analyses indicate the duplication, divergence, and loss of heavy chain and Ca2+-sensors of outer arm dynein among excavate species. These features imply a divergence point with respect to Ca2+-dependent regulation of outer arm dynein in cilia and flagella during the evolution of eukaryotic supergroups.
Similar content being viewed by others
Review
Cilia and flagella are eukaryotic machineries for cell motility propelled by the propagation of bending waves. The internal cytoskeletal structures, called axonemes, are constructed from 9+2 microtubules with axonemal dyneins and regulatory structures such as the central apparatus and radial spokes [1]. These structures are well conserved in all eukaryotes except those that have lost them during evolution. Ciliary and flagellar bend propagations are generated by propagation of sliding of doublet microtubules by axonemal dyneins [2-7]. The propulsive forces generated by bend propagation of cilia and flagella are considered an adaptation for efficient movements by generating fluid flow in microenvironments with low Reynolds numbers [8].
Motility of cilia and flagella is modulated by several extracellular stimuli to enable directed and harmonious movement of cells and tissues. Ca2+ is an important factor for these modulations. Here, I first introduce the diversified roles of Ca2+ in ciliary and flagellar motility over several eukaryotes and then focus on the Ca2+ sensors that directly regulate the motile machinery, the axonemes. In addition, I present a phylogenetic analysis of Ca2+ sensors, demonstrating the evolution of Ca2+ sensors and proposing a pathway of eukaryotic evolution.
Cilia and flagella change their motility in response to Ca2+
Cilia and flagella respond to extracellular stimuli and change their motility. Ca2+ is a well-known intracellular regulator for modulation of ciliary and flagellar movements. These modulations range across diverse modes, including (1) changes in ciliary or flagellar waveforms, (2) rotation or reversal of the direction of ciliary or flagellar bending, (3) arrest of beating, and (4) increasing of beat frequency (Figure 1).
Schematic drawings of various Ca 2+ -dependent changes in wave propagation of cilia and flagella and the direction of locomotion and water flow in several organisms and tissues. Red dots in Ciona sperm and Chlamydomonas flagella indicate acrosomes and mating structure (fertilization tubules), respectively. Black and gray arrows represent the direction of wave propagation and cell locomotion, respectively.
Changes in ciliary or flagellar waveforms
Sperm swim with the tip of the head (acrosome) ahead of the direction of movement for fertilization of the egg. Sperm of the ascidian Ciona intestinalis dramatically increase flagellar asymmetry in response to increases in intracellular Ca2+ concentration caused by a chemoattractant from the egg [9,10]. This change enables the sperm to make turns and move forward towards the egg.
The unicellular alga Chlamydomonas reinhardtii has two flagella and usually swims in breast stroke fashion with the flagella located anterior to the cell body. A structure for mating is formed between the two flagella at fertilization [11,12]. When exposed to intense light, Chlamydomonas stops its motility and then moves in the reverse direction with conversion of flagella into a symmetrical waveform [13]. Analysis with a demembranated cell model suggests that the conversion of flagellar waveform from asymmetric to symmetric is caused by an increase in Ca2+ concentration. The increase of intracellular Ca2+ appears to be performed by Ca2+ influx through a voltage-dependent channel CAV2 [14]. Similar flagellar response to Ca2+ is observed in the prasinophyte Spermatozopsis similis [15].
Rotation or reversal of the direction of ciliary or flagellar bending
In Paramecium, Ca2+ causes reversal of the beating plane of cilia [16-18]. Extracellular stimuli such as mechanical collision induce membrane depolarization and subsequent Ca2+ influx, resulting in ciliary reversal and backward swimming. It is considered that ciliary reversal in Paramecium cilia is caused by the rotation of the central pair in the axoneme [19,20]. Rotation through 180° causes complete reversal of the beating plane of the cilia. In the case of Ctenophora, the ciliary comb plate also shows ciliary reversal in a Ca2+-dependent manner [21]. However, these comb plate cilia perform reversal of the beating plane without rotation of the central axoneme pair [22].
Trypanosoma propagate flagellar waves both from the base to tip and the tip to base [23,24]. Demembranated cell models demonstrate that the direction of flagellar bend propagation reverses when the cell is demembranated by glycerol or detergent and is reactivated by ATP at low concentrations of Ca2+ in the trypanosomatid Crithidia oncopelti [25].
Sperm in some insects and snails reverse the direction of bend propagation in a Ca2+-dependent manner [26-30]. For example, in the sperm of the gastropod Strombus luhuanus, the reversal of bend propagation appears to be involved in sperm release from the sperm storage site in the female genital tract [30].
Arrest of beating
Epithelial cilia of marine invertebrates show ciliary arrest in response to Ca2+. Spontaneous arrest of mussel gill cilia is caused by membrane depolarization, depending on calcium ions [31,32]. Most of the gill cilia in demembranated cell models show arrest of beating at >10−4 M Ca2+ [33]. Ciliary arrest in Ciona stigmatal cells also depends on the presence of external Ca2+ [34]. Cilia of sea urchin embryos or larvae undergo a series of changes in the beating pattern. Spontaneous ciliary arrest is observed in early stages of development; in later stages, cilia show spontaneous reversal or arrest and increase in beat frequency. In many cases, these changes are accelerated by the presence of the Ca2+ ionophore A23187 in seawater [35].
Increase in beat frequency
Ca2+ induces increased beat frequency in airway cilia in mammals [36-38] and in oviductal cilia [39], without alteration of beating direction. Increase in beat frequency is also observed in the Triton-extracted Paramecium model and is inhibited by a calmodulin (CaM) antagonist [18]. However, sperm flagella show no significant increase in beat frequency due to Ca2+ in sea urchin [2] or Ciona (Mizuno and Inaba, unpublished observation), although a demembranated model of sea urchin sperm flagella changed to an asymmetric waveform on stimulation with Ca2+, and showed quiescence at Ca2+ concentration >10−4 M [40].
The effects of Ca2+ on ciliary and flagellar motility appear diverse among organisms, but the roles of Ca2+ in the regulation can be classified into two parts. One is a signaling pathway upstream of the modulation of the axonemes. Influx of Ca2+ is an important trigger for the modulation of ciliary and flagellar motility. Several Ca2+ channels and Ca2+-binding enzymes, such as protein kinases and phosphatases, have been reported to be localized and functional in the ciliary/flagellar plasma membrane and the ciliary/flagellar matrix [6,7]. The other is the direct modulation of axonemal movements. Ca2+-binding proteins such as calaxin, dynein light chain 4 (LC4), CaM, and centrin are bound to the substructures of the axonemes and directly modulate dyneins or their regulatory elements, the radial spokes, and central apparatus. In this paper, I focus on the Ca2+ sensors that directly act on the outer arm dynein in the axonemes.
Outer arm dynein is essential for Ca2+-mediated changes of ciliary movement
The extent of flagellar or ciliary bending correlates with the velocity of microtubule sliding [41,42]. The flagellar waveform is composed of a bend with a larger angle (principal bend) and an opposite bend with a smaller angle (reverse bend) [2]. Formation of bends and propagation are achieved by local microtubule sliding, for which dyneins are considered to be locally activated on one side to bend the axoneme, while those on the other side are inactive [2,43].
The central apparatus (CP) - along with the radial spokes (RS) - plays an important role in flagellar motility as revealed by the paralysis of Chlamydomonas CP mutants [44,45]. The CP is involved in determining the bending plane, demonstrated by the helical movement with 9+0 axonemal structures of eel and Asian horseshoe crab sperm [46,47], and the loss of planar bend movement and the development of helical movement after treatment of a sperm model by antibodies against radial spokes [48]. The activation of specific axonemal dyneins by CP/RS is thought to enable mutual sliding of microtubules across the axoneme, resulting in planar bend propagation [49-51]. Studies on Chlamydomonas flagella have shown that signals from the central apparatus activate specific dyneins for local bending [45,52]. As previously reported, the f (I1) inner arm dynein is regulated by phosphorylation/dephosphorylation of a 138 kDa intermediate chain (IC) through a kinase/phosphatase system present in the RS and CP [53,54].
Axonemes have two dynein motors with different properties: outer arm dynein and inner arm dynein. Subunits of the outer arm dynein have been well studied in Chlamydomonas and in the sperm of Ciona and sea urchins [7,55-59]. They have two or three motor subunits (heavy chains) in the sperm or Chlamydomonas, respectively. Other subunits, including intermediate chains and light chains, are involved in the assembly and regulation of dyneins. Several studies with Chlamydomonas mutants and outer arm extracted sea urchin sperm indicate that outer and inner arm dyneins are involved in the elevation of microtubule sliding velocity (increasing beat frequency) and formation and propagation of flagellar bending, respectively [3,4].
Much experimental evidence demonstrates that outer arm dynein is essential for Ca2+-dependent modulation of ciliary motility. The conversion of flagellar wavelength from symmetric to asymmetric is transiently observed during chemotaxis of the sperm to the egg [9,60,61]. This is caused by Ca2+-dependent regulation of outer arm dynein (see below). Lack of outer arm dynein in the human sperm causes low swimming velocity, loss of circular movement with an asymmetric waveform, and low efficiency of penetration into the egg coat [62,63].
Chlamydomonas changes swimming direction in response to light. There are two types of response: a photophobic reaction to very strong light, photoshock, and a positively or negatively directed movement towards a light source, phototaxis. Both photoshock and phototaxis depend on changes in intracellular Ca2+. Reactivated Chlamydomonas axonemes show an asymmetric beat pattern at Ca2+ concentrations below 10−6 M, become quiescent at 10−5 M, and then resume beating with a symmetric waveform at 10−4 M [64]. This waveform conversion does not occur in mutants lacking dynein outer arms [58,59,65]. In contrast, phototaxis is caused by different responses of the cis- and trans-flagellum. The cis- and trans- flagellar axonemes of demembranated Chlamydomonas cell models differentially respond to Ca2+ concentration in the range 10−8 M to 10−6 M [57]. Studies using axonemal dynein mutants indicate that phototaxis requires the inner, but not the outer, row of dynein arms [58,59].
Specific knockdown of outer arm dynein LC1 in Trypanosoma brucei results in the loss of tip to base propulsive propagation of the flagellar wave [66] that is usually observed in normal forward swimming. A similar phenotype is obtained when LC2 is knocked down [67]. The tip to base propagation is Ca2+-dependent, and the base to tip propagation is only observed in demembranated models when demembranated and reactivated in the presence of EGTA [25]. RNAi knockdown of LC1 in the planarian Schmidtea mediterranea demonstrated that outer arm dynein is essential for the increase of beat frequency and coordination of cilia to produce ciliary oscillation with metachronal waves [68].
Calaxin is the calcium sensor of outer arm dynein necessary for chemotactic turns of the sperm with asymmetric waveforms
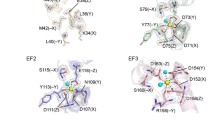
Changes in ciliary and flagellar motility by Ca2+ are mediated by Ca2+-binding proteins. The most common motif for Ca2+ binding is the EF hand. It is a helix-loop-helix structural motif of 12 residues (+X)x(+Y)x(+Z)x(−Y)x(−X)xx(−Z) for metal coordination, where +X, +Y, +Z and −X, −Y, −Z are the vertices of an octahedron [69-71]. The EF hand family contains the CTER, CRP, and S100 subfamilies. These three show mutual congruence to one another within a subfamily. There are many other subfamilies containing EF hands with no strong congruence to one another (Table 1) [72]. Both CTER and CRP basically contain four EF hands, at least one of which lacks the capacity to bind Ca2+ in CRP and does not match the consensus sequence in a PROSITE search (Figure 2A). CTER subfamily proteins, such as CaM, centrin, and troponin C, have dumbbell shape structures with two globular lobes connected by an eight-turn α-helix, whereas CRP, such as recoverin and NCS-1 (frequenin), have a globular structure without the long α-helix link (Figure 2B) [73].
Structures of EF-hand Ca 2+ -binding proteins. (A) Domain structures of Ciona and Chlamydomonas Ca2+-sensors, drawn based on SMART searches (http://smart.embl-heidelberg.de/). The length of each protein and the positions of EF hand motifs are scaled below. (B) Molecular models of ligand-unbound Ciona centrin and NCS-1, built using SWISS-MODEL (http://swissmodel.expasy.org) [175]. Templates used are 1tnx.1 (skeletal muscle troponin) and 2d8n.1 (human recoverin) for Ciona centrin and NCS-1, respectively.
Many studies showed that CaM is an important Ca2+ sensor for the regulation of ciliary and flagellar movements [74,75]. Although CaM was a strong candidate to be the Ca2+-dependent regulator for outer arm dynein in the sperm, several experiments suggest the presence of Ca2+-binding proteins other than CaM. Unlike the light chain 4 (LC4) in Chlamydomonas, the outer dynein could not be isolated from sperm flagella in association with any Ca2+-binding proteins. Moreover, conversion into an asymmetric flagellar waveform is achieved at high concentrations of Ca2+ in the sea urchin sperm model demembranated by Triton X-100 in the presence of millimolar Ca2+ [2,40]. In this condition, CaM is extracted from the axonemes. These reactivated sperm models called ‘potentially symmetric’ sperm show symmetric waveforms at low concentrations of Ca2+ but become asymmetric when Ca2+ is increased in the reactivation medium. The asymmetric flagellar waveform is seen only in the presence of high concentrations of ATP [40], which induces motility with high beat frequency and therefore implies a role of outer arm dynein.
Ca2+-dependent conversion of flagellar waveform is essential for sperm chemotaxis [9,10,60,76-79] and rheotaxis [80], response of sea urchin sperm to mechanical stimuli [81], self-nonself recognition of sperm [82], hyperactivation [83,84], and release from the epithelium of sperm storage sites [85,86]. In the ascidian Ciona intestinalis, correlation between the increase in intracellular Ca2+ concentration and conversion of flagellar asymmetry is clearly observed [9]. Ciona sperm show rather planar wave propagation in seawater with a slight asymmetric flagellar waveform, resulting in a circular trajectory. The reception of the gradient of chemoattractant (sperm activating and attracting factor; SAAF) from the egg [87] induced a transient increase in intracellular Ca2+ concentration. Flagellar axonemes respond to the change and temporarily form and propagate an asymmetric waveform, resulting in a turning movement towards the egg [9].
A previous study found a Ca2+-binding protein that is expressed in Ciona testis during the course of extensive descriptions of axonemal proteins [88]. It turned out that this protein is an axonemal protein localized at the outer arm dynein, named Ca2+-binding axonemal protein calaxin [89]. Calaxin is grouped into one of the CRP EF hand protein families, the neuronal calcium sensor (NCS) protein family, which is expressed in retinal photoreceptors or neurons and neuroendocrine cells [90,91]. A phylogenetic analysis shows that calaxin is a new type of NCS protein in the axoneme; other proteins, such as CaM and centrin, are all grouped into different phylogenetic clades (Figure 3A).
Calaxin is an opisthokont-specific Ca 2+ sensor. (A) A phylogenetic tree of Ca2+-binding proteins in the ascidian Ciona intestinalis. Proteins were aligned by CLUSTALW, and the tree was constructed by MEGA5. Ciona parvalbumin-like protein (XP_002129217) was used as the outgroup. The value shown on each branch represents the number of times that a node was supported in 1,000 bootstrap pseudo-replications. Accession numbers or NCBI reference sequence numbers of the sequence resources are as follows: calmodulin (AB076905), calaxin (AB079059), centrin (XP_004227465), troponin C (XP_002129347), NCS-1 (XP_002126443), hippocalcin (XP_002124848), KChIP (XP_004226075), calcineurin B subunit (CNB) (XP_002130765). (B) Multiple alignment of calaxin in opisthokont species. Asterisks, colons, or dots indicate identical residues in all sequences in the alignment, conserved substitutions, or semi-conserved substitutions, respectively. The amino acid residues identical to Ciona calaxin or to calaxin in other organisms are in red or blue, respectively. The sources of amino acid sequences are as follows: human calaxin (NP_078869), mouse calaxin (NP_080045), Ciona calaxin (AB079059), oyster calaxin (EKC38288), sponge calaxin (XP_003383675), and chytrid fungus calaxin (XP_006677085).
Calaxin has three Ca2+-binding EF hand motifs (amino acids 62 to 90, 98 to 126, and 151 to 166 in Ciona calaxin) [10,89]. Ca2+-binding to these sites was directly demonstrated by isothermal titration calorimetry (ITC), showing a three-site sequential binding model [10]. Two of the three EF hand motifs exhibited endothermic binding and the other exothermic binding. Ca2+-dependent hydrophobic interactions are suggested from positive enthalpy in ITC, as in the case of Ca2+ binding to calmodulin [92]. Several investigations demonstrate membrane-associated roles of NCS in the modulation of neurotransmitter release, biosynthesis of polyphosphoinositides, and in the direct regulation of ion channels [93,94]. In fact, the N-termini of NCS proteins are myristoylated and become exposed outside the protein molecules by binding of Ca2+, allowing them to associate with membranes. The consensus sequence for myristoylation, N-terminal GXXXSX [94], is found in mammalian NCS-1 and calcineurin B. However, it is not present in calaxin or its mammalian orthologs [89], suggesting that the N-terminal is not myristoylated and that calaxin does not have the Ca2+-myristoyl switch property of NCS. Immunohistochemical observations indicate that calaxin is located on the outer arm dyneins along the axoneme of sperm flagella [89]. Calaxin is also distributed in the cilia of ciliated tissues, such as the branchial basket and endostyle [84]. Far western blotting shows that calaxin binds to β-tubulin in the absence of Ca2+ and to the β heavy chain (ortholog of the Chlamydomonas γ heavy chain) of the outer arm dynein [89]a. Calaxin binds to the N-terminal stem region, as revealed by far-western blotting against UV-cleaved fragments of the β heavy chain (Mizuno and Inaba, unpublished data). Although two IQ consensus motifs for binding CaM-like proteins are located within the stem domain of the Chlamydomonas γ heavy chain [95], there is no such motif in the corresponding region of the Ciona β heavy chain.
Ciona sperm shows a unique turning movement associated with a flagellar change to asymmetric waveforms, followed by straight-ahead movement towards the chemoattractant SAAF [87]. In the presence of an NCS inhibitor, repaglinide, the sperm do not exhibit this unique turning movement, showing less-effective chemotaxis [10]. Repaglinide-treated sperm can transiently form asymmetric flagellar waveforms in the gradient of chemoattractant. However, they do not sustain the asymmetric waveform and rapidly return to a symmetric form, resulting in less chemotactic behavior. The flagellar waveforms of sperm demembranated with 0.04% Triton X-100 become more asymmetric when reactivated at >10−6 M Ca2+. Repaglinide attenuates propagation of asymmetric waveforms, but not the relatively symmetric waveforms seen at low concentrations of Ca2+. Calaxin directly suppresses the velocity of microtubule sliding by outer arm dynein at high Ca2+ concentrations. Repaglinide and anti-calaxin antibody cancel the suppression of microtubule translocation at high concentrations of Ca2+. All of these data demonstrate that calaxin plays an essential role in the propagation of asymmetric flagellar bending by suppression of dynein-driven microtubule sliding at high concentration of Ca2+ [10]. Calaxin appears evenly located to every doublet microtubule [89]. Then, how does calaxin work to propagate an asymmetric planar waveform, in which dyneins on the two sides of axonemes mainly participate in microtubule sliding? Although there has not been any experimental evidence to elucidate this question, the function of calaxin might be regulated through a mechanical feedback, such as thrust from flagellar bending, or through a biochemical mechanism, such as protein phosphorylation and dephosphorylation.
BLASTP searches for Ciona calaxin in the genomes of Chlamydomonas reinhardtii and Paramecium tetraurelia hit hypothetical proteins CHLREDRAFT_119565 (XP_001696107) (E = 4e−13) and XP_001433234 (E = 2e−15), respectively. Both hypothetical proteins show a best match with calcineurin subunit B type 1-like protein (CBL-1), not calaxin, in the Ciona genome. LC4 is a Ca2+-binding subunit of outer arm dynein first identified in Chlamydomonas [96]. It shows sequence similarity to CaM and CaM-related proteins such as centrin/caltractin and troponin C. Ca2+-binding assays demonstrate that LC4 has at least one functional Ca2+-binding site. LC4 is isolated in association with the γ heavy chain of outer arm dynein. These properties suggest functions of LC4 analogous to those of calaxin, although the proteins are phylogenetically distinct from each other.
Calaxin is an opisthokont-innovated calcium sensor in cilia and flagella
The current view of eukaryote phylogeny includes its basal division into unikonts (Opisthokonts and Amoebozoa) and bikonts (Archaeplastida, Hacrobia, Stramenopiles, Alveolates, Rhizaria, and Excavata), based on the concept of eukaryotic cells with a single flagellum or two flagella, respectively. Opisthokonts are groups shown to propel cells by a posterior flagellum [97-99]. Homologs of calaxin were searched in available genome databases. Calaxin homologs were not found in any bikont species, such as Archaeplastida (Chlamydomonas) or Stramenopiles (ciliates, dinoflagellates, and blown algae). Calaxin homologs were only found, and were well conserved, in species of the opisthokont supergroup, including Homo sapiens, Mus musculus, Ciona intestinalis, Strongylocentrotus purpuratus, Amphimedon queenslandica, Drosophila melanogaster, Monosiga brevicollis, and Crassostrea gigas. The opisthokont organisms that lack motile cilia or flagella throughout their life cycles, such as C. elegans, Vericrustaceans (except Notostraca and Thecostraca), yeast, and higher fungi show no calaxin gene in their genomes, although these organisms have genes for other NCSs such as NCS-1 (frequenin). The chytrid fungus Batrachochytrium dendrobatidis, grouped into the opisthokonta with metazoa, contains a calaxin gene (XP_006677085) in its genome. The calaxin of B. dendrobatidis shares 38% amino acid identity with Ciona calaxin (Figure 3B). Because of insufficient genome information, the presence of calaxin in Amoebozoa has not been elucidated. BLASTP searches show that calaxin is not present in either the aflagellate amoebozoan Dictyostelium discoideum or the flagellated amoebozoid Breviata anathema which lacks outer arm dynein [100]. However, one of the well-investigated genera in the Amoebozoa, Physarum polycephalum, has a flagellated period in its life cycle. Because it possesses an axoneme of 9+2 structure with outer arm dynein [101-103], it is possible that calaxin could be present in Amoebozoa and could be a unikont-innovated protein.
A previous study identified proteins with a unique combination of domains: an intermediate chain of outer arm dynein, thioredoxin domain and nucleoside diphosphate kinase domain (TNDK-IC, [104,105]) and a radial spoke protein CMUB116 (IQ motif and ubiquitin domain [106]). These proteins are also opisthokont-specific proteins, suggesting that a critical evolutionary event occurred during specification of axonemes in the opisthokont lineage.
Mirror-image relationship between calaxin and LC4
Knowledge of the molecular components of axonemal dyneins and the molecular mechanism of ciliary and flagellar motility has been accumulated mostly from metazoan sperm and certain protists such as Chlamydomonas. In the present study, an attempt has been made to biochemically compare the outer arm dynein and its Ca2+ sensor between Ciona sperm flagella and Chlamydomonas flagella and to correlate their functions in the regulation of motility.
The outer arm dynein of Ciona sperm flagella consists of two heavy chains and represents a two-headed structure, but that of Chlamydomonas flagella consists of three heavy chains with a three-headed structure. Each of the two heavy chains of sperm outer arm dynein is known to have distinct properties [107-110]. Sea urchin α heavy chain (an ortholog of Ciona β and Chlamydomonas γ heavy chains) mediates structural and rigor binding to the microtubules [110]. In vitro motility assays indicate that the absence of the Chlamydomonas γ heavy chain increases both microtubule gliding and ATPase activity [111], suggesting that the γ heavy chain suppresses the activities of outer arm dynein.
Ciona calaxin and Chlamydomonas LC4 bind to Ciona β and Chlamydomonas γ heavy chains, respectively [89,112]. However, Ca2+ dependency of the binding is reversed between Ciona and Chlamydomonas (Figure 4). Calaxin binds to intermediate chain 2 (IC2) and β tubulin in the absence of Ca2+ but becomes associated with the β heavy chain at higher concentrations of Ca2+ [89]. The binding of calaxin to the heavy chain results in the suppression of microtubule-gliding activity by outer arm dynein [10]. In the case of Chlamydomonas, LC4 is bound to the γ heavy chain in the absence of Ca2+ but becomes newly tethered to IC1 (an ortholog of Ciona IC2) in the presence of Ca2+ [95,112]. Although the effect of Ca2+ binding to LC4 on dynein-driven microtubule sliding has not been examined in Chlamydomonas, the binding of Ca2+ to LC4 induces activation of ATPase activity of the outer arm dynein in the mutant lacking the α heavy chain [112]. A model has been proposed for Ca2+-dependent regulation of the γ heavy chain; in the absence of Ca2+, LC4 is tightly bound to the γ HC, resulting in inefficient formation of a rigor bond with microtubules. In the presence of high Ca2+, Ca2+-bound LC4 detaches from the IQ region of the γ heavy chain and becomes attached to IC1, resulting in a structural change of the N-terminal stem domain and the activation of motor activity [95].
Mirror image in the function of outer arm dynein Ca 2+ sensors between Ciona and Chlamydomonas . Ciona calaxin binds to the β-heavy chain, suppresses microtubule-sliding and induces propagation of an asymmetric waveform at high concentration of Ca2+. In contrast, Chlamydomonas LC4 binds to the γ-heavy chain, becomes tethered to IC1 and induces propagation of a symmetric waveform at high concentration of Ca2+. Direct evidence for the activation of microtubule-sliding by Chlamydomonas outer arm dynein has not been obtained.
These mirror-image relationships in the effect of Ca2+ on the regulation of outer arm dynein in Ciona and Chlamydomonas are likely to connect with the difference in the changes of flagellar waveforms (Table 2). At high concentrations of intracellular Ca2+, Ciona sperm show asymmetric waveforms whereas Chlamydomonas flagella become symmetric. The molecular mechanisms of Ca2+-dependent regulation of outer arm dynein appear quite similar to each other, but the response to Ca2+ in the conversion of flagellar waveforms is completely reversed. This implies the possibility of an evolutionary event in the functional diversification of cilia and flagella at the onset of eukaryotic radiation.
It is unlikely that ciliary response in waveform conversion depends on the extracellular Ca2+ concentration in the environment (such as in seawater or freshwater). For example, sperm of freshwater fish show asymmetric waveforms depending on an increase in intracellular Ca2+ concentration [113,114]. The marine alga Pyramimonas parkae shows waveform conversion similar to Chlamydomonas reinhardtii [115], although the relationship between the conversion and intracellular Ca2+ concentration has not been elucidated. An interesting experiment was the examination of the relationship between intracellular Ca2+ concentration and flagellar waveform in the prasinophyte algae Pterosperma and Cymbomonas, both of which show conversion of flagellar waveforms similar to metazoan sperm: symmetric flagellar waveforms in normal swimming and asymmetric waveforms when they change swimming direction [115]. Anterior flagella of Stramenopiles bear hair-like structures called mastigonemes [116]. These organisms or their gametes normally swim with the anterior flagellum ahead. The flagella show symmetric wave propagation from base to tip, but the direction of propulsive force changes because of the reversal of water current by mastigonemes [117]. They change swimming direction in phototactic behavior by altering the flagellar waveform or the orientation of the anterior or posterior flagellum [118], but the relationship between waveform change and intracellular Ca2+ is unclear.
Usage of distinct Ca2+ sensors in unikont and bikont supergroups
A phylogenetic analysis of Ciona calaxin, CaM, centrin, NCS, calcineurin B-subunit (CN-B), Chlamydomonas LC4, and a Ca2+-binding subunit of outer arm dynein docking complex 3 (DC3) [119,120] using available genome information resulted in distinct distribution of calaxin and LC4/DC3 in the opisthokont and bikont supergroups, respectively (Figure 5). Chlamydomonas LC4 and its orthologs were grouped into a clade different from that of calaxin but were more closely related to calaxin than were CaM or centrin. BLASTP searches of Chlamydomonas LC4 against genomes of bikonts resulted in finding orthologs in flagellated species including ciliates, dinoflagellates, diatoms, brown algae, haptophytes, and cryptophytes. Exceptions are seen in organisms lacking outer arm dynein such as angiosperm, moss, and fern [121]. BLASTP searches of Chlamydomonas LC4 against these species resulted in best hits to CaM. Search of Chlamydomonas LC4 in the genomes of opisthokonts failed to find any homologs in this supergroup. For example, the protein most homologous to LC4 in Ciona intestinalis was CaM (E = 3e−22).
Phylogenetic analysis of Ca 2+ -binding proteins. Proteins were aligned by CLUSTALW, and the tree was constructed by MEGA5. Ciona parvalbumin-like protein (XP_002129217) was used as the outgroup. The value shown on each branch represents the number of times that a node was supported in 1,000 bootstrap pseudo-replications. Sequences were obtained from the organisms Ciona (Ciona intestinalis), human (Homo sapiens), fungus (Batrachochytrium dendrobatidis), Naegleria (Naegleria gruberi), Euglena (Euglena gracilis), Trypanosoma (Trypanosoma cruzi or T. brucei), Giardia (Giardia intestinalis or G. lamblia), Trichomonas (Trichomonas vaginalis), Chlamydomonas (Chlamydomonas reinhardtii), Paramecium (Paramecium tetraurelia), and Ectocarpus (Ectocarpus siliculosus). The sources of amino acid sequences are as follows: Ciona calmodulin (AB076905), Ciona calaxin (AB079059), Ciona centrin (XP_004227465), Ciona NCS-1 (XP_002126443), Ciona CNB (XP_002130765); human CaM (CAA36839), human calaxin (NP_078869), human NCS1 (NP_055101), human CNB (NP_000936), human centrin (NP_004057); chytrid fungus calaxin (XP_006677085), chytrid fungus CaM (XP_006678916), chytrid fungus centrin (XP_006682970), chytrid fungus NCS1 (XP_006675998), chytrid fungus CNB (XP_006677028); Naegleria CaM (XP_002683533), Naegleria centrin (XP_002678269); Trypanosoma CaM (XP_805243), Trypanosoma centrin (XP_805423), Trypanosoma calflagin (Q26680); Euglena CaM (P11118), Euglena centrin (AGS09408); Giardia CaM (XP_001705820), Giardia centrin (XP_001707577), Giardia LC4 (XP_001705117); Trichomonas CaM (XP_001326924), Trichomonas centrin (CAB55607), Trichomonas CNB (XP_002680632); Paramecium CaM (XP_001448363), Paramecium LC4 (XP_001442002), Paramecium centrin (XP_001347281), Paramecium DC3 (XP_001444482); Ectocarpus LC4 (CBN80105), Ectocarpus CaM (CBN74265), Ectocarpus centrin (CBN79657), Ectocarpus DC3 (CBJ30770). The protein sequences with specific accession numbers were obtained from DDBJ/EMBL/GenBank, or from genome browsers with the following URLs: Chlamydomonas http://genome.jgi-psf.org/Chlre4/Chlre4.home.html; Paramecium http://paramecium.cgm.cnrs-gif.fr; Naegleria http://genome.jgi-psf.org/Naegr1/Naegr1.home.html; Trichomonas http://trichdb.org; and Trypanosoma https://www.sanger.ac.uk/resources/downloads/protozoa/trypanosoma-brucei.html.
DC3 is also a CaM type of EF hand protein localized on the outer arm dynein docking complex and shows redox-sensitive Ca2+-binding with a ratio of 1 mol Ca2+/mol protein [120]. However, it is unclear whether DC3 actually binds Ca2+ under physiological conditions because it also significantly binds Mg2+ [122]. Genes of DC3 homologs are present in Bikonta such as Stramenopiles (ciliates, brown algae, and Plasmodium) and Cryptophytes but could not be found in the Ciona or human genomes. DC3 grouped into a clade closer than LC4 to CNB/calaxin/NCS (Figure 5). Intriguingly, BLASTP searches using recent genomic information on the chlorarachniophyte Bigelowiella natans did not detect orthologs of Chlamydomonas LC4 or DC3. The protein with highest similarity was CaM (ID 54077), although ultrastructural observation of the flagella clearly shows the presence of outer arm dynein [123]. LC4 was also absent from Plasmodium (Apicomplexa).
Both CN-B and NCS have been found in animals and fungi [124] but do not appear in plants. In plants, the CNB-like protein (CBL) family represents a unique group of calcium sensors and plays a key role in intracellular Ca2+ signaling [124]. CNB-like proteins in plants are most closely related to CNB and NCS proteins in animals and fungi (Figure 5). Proteins in Chlamydomonas (ID391130) and in Paramecium (GSPATP9660001) are grouped with CNB-like protein. Separation of these proteins from the CNB group is supported by the bootstrap value (986/1,000).
Phylogenetic analysis of homologs of Ca 2+ sensor proteins in Excavata. Proteins (EF-hand proteins, length less than 350 amino acids) were searched against genomes of each excavate by BLASTP and those with E-value <e−9 were aligned with Ciona or Chlamydomonas Ca2+-sensors by CLUSTALW. An unrooted tree was drawn by MEGA5. Branches of each Ca2+-sensor are highlighted by colors. The protein sequences (with accession numbers indicated) were obtained from DDBJ/EMBL/GenBank, or from the genome browsers shown in the legend of Figure 5.
The supergroup Excavata includes eight taxa [125-128]. Phylogenetic analysis supports the monophyly of the Excavata [128] which consists of two major groups, Discoba and Metamonada. An additional organism, Malawimonas, may also be included as a genus in the Excavata. Discoba includes four phyla, Jakobida, Euglenozoa (for example, Euglena, Trypanosoma), Heterolobosea (for example, Naegleria), and Tsukubamonadida. The Metamonada includes amitochondriate flagellate Fornicata (for example, Giardia), Parabasalids (for example, Trichomonas), and Preaxostyla [126]. Although Excavata are often considered the extant organisms closest to the ancient eukaryotes, there are debates concerning their phylogenetic position.
Analysis of Ca2+ sensors in Excavata leads to an interesting point of view concerning the evolution of Ca2+ sensor proteins (Figures 5, 6 and 7). First, both Giardia lamblia (XP_001705117) and Naegleria gruberi (ID 70962) contain clear orthologs of Chlamydomonas LC4 (Figure 5). Second, Naegleria has clear orthologs of NCS-1 and CNB (Figure 5). Third, several excavate species have multiple proteins with similarity to CNB, NCS-1, LC4, or DC3 (Figure 6), although they could only be grouped into each Ca2+ sensor family with weak bootstrap support. Euglena has three DC3-like proteins. Naegleria has a LC4-like protein. Trypanosoma Tb10707970 is a CNB-like protein. Trichomonas has three NCS-1-like proteins. There are other proteins in Trichomonas, Naegleria, and Euglena that are similar to, but could not be grouped with, any ciliary Ca2+ sensors (Figures 6 and 7). These features of Ca2+ sensors or their homologs in Excavata suggest that duplication and divergence of Ca2+ sensors occurred in this supergroup.
Distribution of Ca 2+ sensor proteins in eukaryotes. Based on the BLASTP search and the phylogenetic analyses in Figures 5 and 6, occurrence of each Ca2+ sensor in eukaryotic groups is summarized. Occurrence is indicated in the same colors as used in Figures 5 and 6. Closed circles in a specific color represent an occurrence of homologs with weak bootstrap support.
Structure of outer arm dynein and its Ca 2+ sensor across eukaryotic groups. (A) Schematic representation of the number of dynein heavy chains and the morphology of outer arm dyneins observed by electron microscopy. Chlamydomonas outer arm dynein is composed of three heavy chains, α, β, and γ. Ciona outer arm dynein has two heavy chains homologous to the Chlamydomonas β and γ chains. The α and β heavy chains in Ciona and the β and α heavy chains in sea urchin correspond to Chlamydomonas β and γ, respectively. ODA, outer arm dynein; IDA, inner arm dynein; N-DRC, nexin link/dynein regulatory complex. (B) Distribution of two-headed or three-headed outer arm dynein, and calaxin or LC4, across eukaryotic groups. The occurrence of calaxin or LC4 is indicated in red or blue, respectively, in the name of the group. A group name in black or gray indicates the lack of both calaxin and LC4, or not enough genomic information, respectively. The references for the EM images of the axonemes and the outer arm dynein are as follows: Naegleria [146]; Euglena [176,177]; Trypanosoma [66,67]; Giardia [144]; Trichomonas [147]: amoebozoan (Physarum) [101-103]; choanoflagellate (Codosiga botrytis) [178]; chordate (Ciona intestinalis and human) [62,88]; echinoderm (sea urchin: Colobocentrotus atratus) [1,3]; platyhelminthes (Dugesia tigrina) [68,179]; arthropod (Exechia seriara) [180]; Mollusca (Crassostrea gigas) [181]; chytrid fungus (Rhizophlyctis) [182]; green alga (Chlamydomonas) [137]; diatom (Biddulphia levis) [183]; golden alga (Ochromonas) [116]; ciliate (Tetrahymena pyriformis) [184]; dinoflagellate (Wolszymkia micra) [185]; apicomplexan (Plasmodium) [141]; chlorarachnion (Bigelowiella natans) [123]; haptophyte (Chrysochromulina) [186]; and phytomyxean (Plasmodiophora brassicae) [140].
Ca2+ sensors appear to evolve with dynein heavy chains
As described above, Ciona and Chlamydomonas use distinct Ca2+ sensors for outer arm dynein. The molecular properties of these two proteins differ from each other, and this might be related to the difference in Ca2+-dependent regulation of flagellar motility. BLAST searches using genomic information from several organisms indicate that calaxin is an opisthokont-specific protein. Orthologs of Chlamydomonas LC4 are distributed in Archaeplastida, Alveolata, Stramenopiles, Cryptophytes, Giardia and Naegleria, but not in Opisthokonta or the excavates Euglena and Trypanosoma.
Ca2+ sensors directly act on the motor subunits of outer arm dynein. The heavy chains of outer arm dynein are phylogenetically classified into ODAα and ODAβ families [129]. The ODAα family includes the Chlamydomonas γ heavy chain, the Ciona β heavy chain, and the sea urchin α heavy chain, all of which are located at the innermost part of the outer arm [130,131]. The ODAβ family includes the Chlamydomonas α and β heavy chains, the Ciona α heavy chain, and the sea urchin β heavy chaina.
It is known that the number of heavy chains of outer arm dynein is two in metazoan sperm but three in Chlamydomonas and ciliates [132-136]; from the molecular structure of dynein, they are called two-headed and three-headed. EM images of cross sections of the axonemes enable analysis of the number of heavy chains of outer arm dynein (Figure 8A; [133]). The outer arm of a Chlamydomonas mutant lacking the α heavy chain lacks the outermost part and appears similar to the outer arm of sperm flagella [137,138], indicating that the outermost part corresponds to the α heavy chain. Other observations by transmission electron microscopy (TEM) [138] or cryo-electron tomography [130,131] indicate that the innermost part and the center part of the TEM image is composed of the γ and β heavy chain in Chlamydomonas, respectively. Following the idea of Mohri et al. [133], the number of heavy chains could be predicted from the morphology of outer arm dynein observed by TEM (Figure 8A). I surveyed published TEM images of outer arm dyneins in several organisms. It is intriguing to note that the number of dynein heads and the Ca2+ sensor used for regulation of outer arm dynein turn out to be well correlated (Figure 8B).
A possible model for the evolution of, and diversification in, the structures of outer arm dynein and corresponding Ca 2+ sensors during eukaryotic evolution. The model is based on analyses of the structures of outer arm dynein (two-headed, three-headed) and the types of Ca2+-sensor in each group of eukaryotes. It is assumed that the heavy chains and Ca2+-sensors of outer arm dynein of the last eukaryotic common ancestor (LECA) preceded duplication, and that duplication and divergence of Ca2+-sensors occurred at an early stage of eukaryotic diversification. The model is arranged so that the positions of eukaryotic groups match with widely accepted phylogenetic relationships [128,158]. The number of cilia/flagella per cell is also indicated in parenthesis (brown letters). Note that the numbers of cilia/flagella in Euglena and Trypanosoma are indicated as ‘1+,’ since these organisms are considered to have been biflagellates but lost or largely degenerated one of the two flagella during evolution. In this model, duplication of dynein heavy chain occurred at the root of the bikont lineage. Duplication and divergence of Ca2+-sensors would have already occurred in the ancestral organisms that contained three-headed dynein. An ancestral organism containing three-headed dynein might have recruited LC4-like sensors or CNB/NCS-like sensors and then branched into the Metamonadan (Trichomonas + Giardia) and Discoban lineages. Loss of dynein heavy chains would have occurred in Giardia and the Euglenozoa. Red or blue asterisks represent duplication or loss of a dynein heavy chain, respectively. Colored dots next to the two- or three-headed dyneins represent Ca2+-sensors (red, calaxin; blue, LC4; magenta, DC3; green, NCS; cyan, CNB). In the lineage of opisthokonts or Archaeplastida/Stramenopile/Alveolata, calaxin, LC4 or DC3 is demonstrated to be bound to the dynein heavy chain, although it is not known whether Ca2+-sensors in Excavates or any of the hypothetical ancestors could bind to the dynein or not.
It is believed that the two heavy chains of the ODAβ family resulted from gene duplication [139], but the exact phylogenetic position of the duplication is not clear. The biflagellated swarm cells in the amoebozoan Physarum possess 9+2-structured flagella. Cross sections of Physarum axonemes suggest that the outer arm dynein is two-headed [101-103], like those in opisthokonts. However, the presence of calaxin and the number of heavy chains in the outer arm dynein remain unclear because of the lack of a genome sequence. Recent genome information reveals no gene similar to Chlamydomonas LC4 or DC3 in the chlorarachnion Bigelowiella natans. The number of heavy chains is possibly three judged from an EM image [123]. Another cercozoan, Plasmodiophora brassicae, apparently possesses three-headed outer arm dynein [140], but no genomic information is available. Ciliates, such as Paramecium and Tetrahymena, have three-headed outer arm dynein and a gene orthologous to Chlamydomonas LC4. However, another group of Alveolata, the Apicomplexa, shows a different feature; the axonemes of Plasmodium berghei have normal 9+2 structure with three-headed outer arm dynein [141]. It is not clear whether P. berghei has LC4 since the genome sequence of this organism is not available. The gregarin Lecudina tuzetae has a 6+0 structured axoneme, but the detailed structure of the outer arm dynein is unclear from the available EM images [142].
Six species in the Excavata were available for prediction of the number of heavy chains from EM images. First, the euglenozoan species Euglena, Leishmania, and Trypanosoma show a two-headed shape of outer arm dynein. The genome sequences reveal that neither Euglena nor Trypanosoma have LC4. Second, Giardia has an LC4 homolog in the genome. EM images, however, are very close to those of two-headed outer arm dynein [143,144]. This might be because Giardia lamblia is a fast-evolving parasitic species, leading to an error in phylogenetic analysis due to long-branch attraction (LBA) [145]. Lastly, the outer arm dyneins of two excavate species, Naegleria gruberi and Trichomonas vaginalis, appear three-headed, although little TEM data with clear images of outer arm dynein is available [146,147].
Eukaryote evolution in view of outer arm dynein and its calcium sensors
The structure of the axoneme and the regulation of ciliary and flagellar motility are basic aspects of all major eukaryotic groups and undoubtedly one of the ancestral features of eukaryotes [148-151]. There are three hypotheses for how cilia were acquired in the last eukaryotic common ancestor (LECA): endosymbiosis of a Spirochete and an Archaebacterium [152], viral infection [153], and autogenous origin [153] (see reviews [149,154]). The latter hypothesis is widely accepted at present. During overall evolution of cell motility, ciliary movement and amoeboid movement were selectively or cooperatively used depending on the body plan of the organisms. In the most probable LECA unicellular organism, both ciliary and amoeboid locomotion systems appear to have been used [151]. Ancient flagella are considered to be used for attachment to a substrate and to pull the organism by gliding. It is possible that flagella then acquired regulatory systems for directed, tactical, or avoiding movement with high speed, with the aid of extracellular signaling molecules such Ca2+; examples of such regulated movement are reversal of bend propagation and altering of flagellar waveforms (Figure 1). In this case, as much evidence indicates, Ca2+-dependent regulation of the outer arm dynein is thought to be critical. During diversification, some organisms lost components of the axoneme. For example, loss of outer arm dynein is probably due to the loss of a requirement for rapid and/or extensive reorientation of the cell. Other organisms have lost motile flagella or cilia, probably due to disuse of their motility, in, for example, reproduction. The former include the gregarin Lecudina tuzetae, Breviata, fern, moss, eel, and insects like Acerentomon microrhinus, and the latter include nematodes, crustaceans, and angiosperms [154,155].
Taking into account the fact that cilia have been inherited through the major pathways of eukaryotic evolution, I here propose a hypothesis for eukaryotic evolution based on phylogenetic analyses of Ca2+ sensors and the number of dynein heads. The most evident feature is that the majority of opisthokonts show two-headed outer arm dynein with the Ca2+ sensor calaxin, whereas the majority of bikonts (Archaeplastida, Stramenopiles, Alveolata, and some (but not all) Excavata) have three-headed outer arm dynein with Chlamydomonas LC4-type Ca2+ sensors. Excavata robustly emerge between unikonts and Archaeplastida/Hacrobia/Stramenopiles/Alveolata/Rhizaria and form a monophyletic supergroup [128]. Several phylogenetic analyses of diverse eukaryotes have led to the idea that the eukaryotic root could be set at the base between unikonts and bikonts [156-158], but this is still controversial [158-162].
The Excavata is certainly a supergroup that could provide key clues to understanding the evolution of dynein and its Ca2+ sensors and shed light on the origin of Ca2+-dependent regulation of cilia and flagella. A phylogenetic analysis in this study showed that excavates had already evolved several Ca2+ sensors, including those with similarities to extant Ca2+ sensors. Based on the widely accepted relationship among excavate species [128,158], a possible pathway could be considered with respect to evolution of dynein structure and Ca2+ sensors (Figure 9). This model is based on the hypothesis that the LECA had two-headed dynein and that Ca2+ sensors were duplicated in the initial stage of eukaryotic evolution and became divergent (and then possibly became functional) during evolution. Loss of dynein heavy chains or Ca2+ sensors in Excavata, possibly by reduction of genomes in obligate parasites [143,163], is also taken into consideration.
The duplication of dynein heavy chains would have occurred at the root of the bikont lineage (Figure 9). From the strong bootstrap supports (Figure 5), it appears that three-headed dynein might have recruited LC4 in the last common ancestor of bikonts, which would be involved in the diversification in Metamonada (Trichomonas and Giardia). Similarly, CNB/NCS-like Ca2+ sensor homologs must have existed in the last common eukaryotic ancestor. Another route for Discoba diversification might have involved retentions of CNB/NCS-like Ca2+ sensors.
Excavates show a variety in the number of motile flagella per cell. For example, the euglenoids Trypanosoma brucei and Euglena gracilis are biflagellate but one of the two flagella is highly reduced. There are two flagella in Naegleria gruberi, five flagella in Trichomonas vaginalis, and eight flagella in Giardia lamblia (see Figure 9). It is worth pointing out that the excavate species bearing a single motile flagellum, that is, Euglena and Trypanosoma, have two-headed dyneins; Giardia is the only excavate with two-headed dynein and multiple flagella (Figure 9). The only other eukaryotic group containing organisms (or cells) with a single motile flagellum is the Opisthokonta.
The Amoebozoa, Physarum polycephalum and Breviata anathema, originally grouped into unikonts [125], bear two basal bodies. It has therefore been debated whether Amoebozoa and Opisthokonta can be monophyletically grouped [157,164]. Physarum has one long and one short flagellum connected to two basal bodies, and Breviata anathema, a small amoeba-like cell, has a single flagellum from each of the two basal bodies. The presence of two basal bodies is proposed as one of the characteristics of bikonts [165]. From TEM images of axonemes, Physarum appears to have two-headed outer arm dyneins (Figure 8), which is a common aspect of opisthokonts [133]. Breviata does not have outer arm dynein [100], meaning there is no evidence for its grouping based on the criterion of the structure of outer arm dynein. It would be intriguing to search for calaxin (also TNDK-IC and CMUB, see above and [166]) in organisms that have been under debate in terms of classification into bikonts or unikonts.
New genes with novel functions are evolved by gene duplication [167]. Several models have been proposed for mechanisms of how new protein functions evolve through gene duplication and divergence [168]. Recruitment of functional Ca2+ sensors seems particularly important in cilia and flagella because they participate in gamete motility, essential for the success of reproduction in most organisms. For Ca2+ sensors of outer arm dynein, the functions of calaxin and Chlamydomonas LC4 regulate the motor activity in flagella, but their response to Ca2+ concentration is different. The distribution of these Ca2+ sensors in extant species in eukaryotes is described in the present paper. Calaxin and LC4 appear to be preserved in Opisthokonta and the majority of bikonts (Archaeplastida, Stramenopiles, and Alveolata), respectively.
It is possible that these proteins became preserved after protein evolution by gene duplication and divergence because of their specific functions in the interaction with the cytoskeleton and the regulation of a molecular motor. The module-dominant conservation, as seen in axonemes [166], is possibly because of the need for preservation of multiple proteins in this cytoskeletal architecture. No biochemical evidence has been obtained for the localization or functions of Ca2+ sensors, except Ciona calaxin and Chlamydomonas LC4. To learn whether evolution of proteins by gene duplication and divergence accompanies or precedes innovation of protein function, it would be fascinating to examine the interaction of an ancient calaxin with microtubules or dyneins.
Conclusions
Conversion from asymmetric to symmetric movement at high concentrations of Ca2+ requires outer arm dynein in Chlamydomonas flagella. Conversion to an asymmetric waveform in sperm flagella is also performed by outer arm dynein at high Ca2+ concentration. Thus, the functions of outer arm dynein are regulated by Ca2+ sensors at high concentrations of Ca2+ in both Chlamydomonas and sperm flagella. Recruitment of Ca2+ sensors to outer arm dynein might have made it possible for the organisms to respond to ‘high’ Ca2+ to modulate flagellar waveforms to change their direction of movement, although the directions of conversion of waveforms are a mirror-image of each other in Chlamydomonas and sperm.
In this paper, it is suggested that the duplication and divergence of Ca2+-sensors might have occurred at an early stage in eukaryotic evolution. The clear distinction in dynein structure and Ca2+ sensors between opisthokonts and bikonts, and their heterogeneity in Excavata, suggests an important role of ciliary regulation in eukaryotic evolution. It is unclear, however, if Ca2+ sensors in Excavata really function in the regulation of outer arm dynein. Outer arm dynein in Trypanosoma is essential for tip-to-base movement, which is induced by ‘low’ intracellular Ca2+. Loss of outer arm dynein results in a defect of tip-to-base movement in response to low intracellular Ca2+. This feature of Ca2+ regulation of outer arm dynein is distinct from that observed in Chlamydomonas and Ciona. Trypanosoma and Naegleria have CaM in flagella called flagellar CaM or CaM-1. CaM is localized in paraflagellar rods and regulates their assembly in Trypanosoma [169]. However, it is unclear whether CaM is localized to the outer arm dynein or other axonemal structures. Further studies are necessary to elucidate the role of Ca2+-binding proteins in the regulation of the outer arm dynein in Excavata.
Calaxin was acquired in Opisthokonta and may participate not only in the regulation of fluid flow mediated by cilia and flagella but also in other phenomena that characterize opisthokonts, such as cell polarity, differentiation of nerve cells, and establishment of body plan. The first definition of Opisthokonta by Cavalier-Smith [170], that is, organisms having posterior flagella to propel cells forward, may be related to the position of the sperm acrosome in the anterior part where sperm adhere and fuse with the counterpart gamete egg. The corresponding portion of Chlamydomonas, the mating structure, sits between two flagella. Both Chlamydomonas and sperm move forward with these mating structures at the leading edge (Figure 1). Differentiation of the sperm acrosome accompanies the localization of the Golgi apparatus and vesicles at the anterior of the head [171,172]. The Golgi apparatus and vacuoles are likely to locate near the flagella of Chlamydomonas [173], implying that the intracellular compartments for gamete recognition are reversely positioned relative to the positions of basal bodies between sperm and Chlamydomonas. Although it is not known whether this difference in cell polarity is connected to the mirror-image of Ca2+-dependent regulation between these cells, experiments such as knockout of the calaxin gene in metazoa might give important insights into the evolutionary relationship between cilia and organisms.
The present study implies early events in the diversification of Ca2+ sensors for outer arm dynein during evolution, but connections of the phylogenetic view of outer arm dyneins to the function or motility of cilia and flagella have not been completely clarified. Most of the discussion in this paper is based on the assumption that orthologous proteins conserve their function across species, but this is not always the case. For solving such problems, it is evidently necessary to confirm whether the proteins used in phylogenetic analyses in the present study are localized and bound to the ciliary or flagellar axonemes and function as Ca2+ sensors of outer arm dynein. The precise number of heads must also be determined by observation with cryo-electron tomography.
Abbreviations
- EM:
-
electron microscopy
- IC:
-
intermediate chain
- LC:
-
light chain
- NCS:
-
neuronal calcium sensor
- RNAi:
-
RNA interference
References
Gibbons IR (1981) Cilia and flagella of eukaryotes. J Cell Biol 91:107s–124s
Brokaw CJ (1979) Calcium-induced asymmetrical beating of triton-demembranated sea urchin sperm flagella. J Cell Biol 82:401–411
Gibbons BH, Gibbons IR (1973) The effect of partial extraction of dynein arms on the movement of reactivated sea-urchin sperm. J Cell Sci 13:337–357
Brokaw CL, Kamiya R (1987) Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton 8:68–75
Lindemann CB (2004) Testing the geometric clutch hypothesis. Biol Cell 96:681–690
Inaba K (2003) Molecular architecture of the sperm flagella: molecules for motility and signaling. Zool Sci 20:1043–1056
Inaba K (2011) Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol Hum Reprod 17:524–538
Sleigh MA (1991) Mechanisms of flagellar propulsion. A biologist’s view of the relation between structure, motion, and fluid mechanics. Protoplasma 164:45–53
Shiba K, Baba SA, Inoue T, Yoshida M (2008) Ca2+ bursts occur around a local minimal concentration of attractant and trigger sperm chemotactic response. Proc Natl Acad Sci USA 105:19312–19317
Mizuno K, Shiba K, Okai M, Takahashi Y, Shitaka Y, Oiwa K, Tanokura M, Inaba K (2012) Calaxin drives sperm chemotaxis by Ca2+-mediated direct modulation of a dynein motor. Proc Natl Acad Sci USA 109:20497–20502
Wilson NF, Foglesong MJ, Snell WJ (1997) The Chlamydomonas mating type plus fertilization tubule, a prototypic cell fusion organelle: isolation, characterization, and in vitro adhesion to mating type minus gametes. J Cell Biol 137:1537–1553
Goodenough U, Lin H, Lee JH (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18:350–361
Witman GB (1993) Chlamydomonas phototaxis. Trends Cell Biol 3:403–408
Fujiu K, Nakayama Y, Iida H, Sokabe M, Yoshimura K (2011) Mechanoreception in motile flagella of Chlamydomonas. Nat Cell Biol 13:630–632
McFadden GI, Schulze D, Surek B, Salisbury JL, Melkonian M (1987) Basal body reorientation mediated by a Ca2+-modulated contractile protein. J Cell Biol 105:903–912
Eckert R, Naitoh Y, Machemer H (1976) Calcium in the bioelectric and motor functions of Paramecium. Syrup Soc Exp Biol 30:233–255
Naitoh Y, Kaneko H (1972) Reactivated Triton-extracted models of Paramecium: modification of ciliary movement by calcium ions. Science 176:523–524
Nakaoka Y, Tanaka H, Oosawa F (1984) Ca2+-dependent regulation of beat frequency of cilia in Paramecium. J Cell Sci 65:223–231
Omoto CK, Kung C (1979) The pair of central tubules rotates during ciliary beat in Paramecium. Nature 279:532–534
Omoto CK, Kung C (1980) Rotation and twist of the central-pair microtubules in the cilia of Paramecium. J Cell Biol 87:33–46
Nakamura S, Tamm SL (1985) Calcium control of ciliary reversal in ionophore-treated and ATP-reactivated comb plates of ctenophores. J Cell Biol 100:1447–1454
Tamm SL, Tamm S (1981) Ciliary reversal without rotation of axonemal structures in ctenophore comb plates. J Cell Biol 89:495–509
Holwill MEJ (1965) The motion of Strigomonas oncopelti. J Exp Biol 43:125–137
Jahn TL, Bovbe EC (1968) Locomotion of blood protists. In: Weinman D, Ristie M (eds) Infectious blood diseases of man and animals. Academic Press, New York, pp 393–436
Holwill ME, McGregor JL (1976) Effects of calcium on flagellar movement in the trypanosome Crithidia oncopelti. J Exp Biol 65:229–342
Baccetti B, Gibbons BH, Gibbons IR (1989) Bidirectional swimming in spermatozoa of Tephritid flies. J Submicrosc Cytol Pathol 21:619–625
Ishijima S, Ishijima SA, Afzelius BA (1999) Movement of turritella spermatozoa: direction of propagation and chirality of flagellar bends. Cell Motil Cytoskeleton 44:85–95
Yang Y, Lu X (2011) Drosophila sperm motility in the reproductive tract. Biol Reprod 84:1005–1015
Köttgen M, Hofherr A, Li W, Chu K, Cook S, Montell C, Watnick T (2011) Drosophila sperm swim backwards in the female reproductive tract and are activated via TRPP2 Ion channels. PLoS ONE 6:1–8
Shiba K, Shibata D, Inaba K (2014) Autonomous changes in the swimming direction of sperm in the gastropod Strombus luhuanus. J Exp Biol 217:986–996
Murakami A, Takahashi K (1975) The role of calcium in the control of ciliary movement in Mytilus. II. The effects of calcium ionophores X537A and A23187 on the lateral gill cilia. J Fac Sci Univ Tokyo IV 13:251–256
Tsuchiya T (1977) Effects of calcium ions on Triton-extracted lamellibranch gill cilia: ciliary arrest response in a model system. Comp Biochem Physiol 56A:353–361
Walter MF, Satir P (1979) Calcium does not inhibit active sliding of microtubules from mussel gill cilia. Nature 278:69–70
Takahashi T, Baba S, Murakami A (1973) The “excitable” cilia of the tunicate, Ciona intestinalis. J Fac Sci Univ Tokyo Sect 1V Zool 13:123–137
Degawa M, Mogami Y, Baba SA (1986) Developmental changes in Ca2+ sensitivity of sea-urchin embryo cilia. Comp Biochem Physiol 82A:83–90
Kakuta Y, Kanno T, Sasaki H, Takishima T (1985) Effect of Ca2+ on the ciliary beat frequency of skinned dog tracheal epithelium. Respir Physiol 60:9–19
Sanderson MJ, Dirksen ER (1986) Mechanosensitivity of cultured ciliated cells from the mammalian respiratory tract: implications for the regulation of mucociliary transport. Proc Natl Acad Sci USA 83:7302–7306
Benedetto GD, Magnus CJ, Gray PT, Mehta A (1991) Calcium regulation of ciliary beat frequency in human respiratory epithelium in vitro. J Physiol 439:103–113
Verdugo P (1980) Ca2+-dependent hormonal stimulation of ciliary activity. Nature 283:764–765
Gibbons BH, Gibbons IR (1980) Calcium-induced quiescence in reactivated sea urchin sperm. J Cell Biol 84:13–27
Brokaw CJ (1972) Flagellar movement: a sliding filament model. Science 178:455–462
Brokaw CJ (1991) Microtubule sliding in swimming sperm flagella: direct and indirect measurements on sea urchin and tunicate spermatozoa. J Cell Biol 114:1201–1215
Sale WS (1986) The axonemalaxis and Ca2+-induced asymmetry of active microtubule sliding in sea urchin sperm tails. J Cell Biol 102:2042–2052
Witman GB, Plummer J, Sander G (1978) Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol 76:729–747
Smith EF, Yang P (2004) The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton 57:8–17
Gibbons BH, Baccetti B, Gibbons IR (1985) Live and reactivated motility in the 9+0 flagellum of Anguilla sperm. Cell Motil 5:333–350
Ishijima S, Sekiguchi K, Hiramoto Y (1988) Comparative study of the beat patterns of American and Asian horseshoe crab sperm: evidence for a role of the central pair complex in forming planar waveforms in flagella. Cell Motil Cytoskeleton 9:264–270
Gingras D, White D, Garin J, Cosson J, Huitorel P, Zingg H, Cibert C, Gagnon C (1998) Molecular cloning and characterization of a radial spoke head protein of sea urchin sperm axonemes: involvement of the protein in the regulation of sperm motility. Mol Biol Cell 9:513–522
Bannai H, Yoshimura M, Takahashi K, Shingyoji C (2000) Calcium regulation of microtubule sliding in reactivated sea urchin sperm flagella. J Cell Sci 113:831–839
Wargo MJ, Smith EF (2003) Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc Natl Acad Sci USA 100:137–142
Mitchell DR (2003) Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil Cytoskeleton 56:120–129
Smith EF, Sale WS (1992) Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science 257:1557–1559
Habermacher G, Sale WS (1997) Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol 136:167–176
King SJ, Dutcher SK (1997) Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol 136:177–1191
King SM (2000) The dynein microtubule motor. Biochim Biophys Acta 1496:60–75
King SM (2010) Sensing the mechanical state of the axoneme and integration of Ca2+ signaling by outer arm dynein. Cytoskeleton 67:207–213
Kamiya R, Witman GB (1984) Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J Cell Biol 98:97–107
Kamiya R, Okamoto M (1985) A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J Cell Sci 74:181–191
Mitchell DR, Rosenbaum JL (1985) A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol 100:1228–1234
Cosson MP, Carré D, Cosson J (1984) Sperm chemotaxis in siphonophores. II. Calcium-dependent asymmetrical movement of spermatozoa induced by the attractant. J Cell Sci 68:163–181
Wood CD, Darszon A, Whitaker M (2003) Speract induces calcium oscillations in the sperm tail. J Cell Biol 161:89–101
Afzelius BA, Eliasson R, Johnsen O, Lindholmer C (1975) Lack of dynein arms in immotile human spermatozoa. J Cell Biol 66:225–232
Wolf JP, Feneux D, Escalier D, Rodrigues D, Frydman R, Jouannet P (1993) Pregnancy after subzonal insemination with spermatozoa lacking outer dynein arms. J Reprod Fertil 97:487–492
Bessen M, Fay RB, Witman GB (1980) Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J Cell Biol 86:446–455
Wakabayashi K, Yagi T, Kamiya R (1997) Ca2+-dependent waveform conversion in the flagellar axoneme of Chlamydomonas mutants lacking the central-pair/radial spoke system. Cell Motil Cytoskeleton 38:22–28
Baron DM, Kabututu ZP, Hill KL (2007) Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J Cell Sci 120:1513–1520
Branche C, Kohl L, Toutirais G, Buisson J, Cosson J, Bastin P (2006) Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci 119:3443–3455
Rompolas P, Patel-King RS, King SM (2010) An outer arm dynein conformational switch is required for metachronal synchrony of motile cilia in planaria. Mol Biol Cell 21:3669–3679
Kretsinger RH, Nockolds CE (1973) Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem 248:3313–3326
Kawasaki H, Nakayama S, Kretsinger RH (1998) Classification and evolution of EF-hand proteins. Biometals 11:277–295
Grabarek Z (2006) Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol 359:509–525
Permyakov EA, Kretsinger RH (2011) Calcium binding proteins, John Wiley & Sons, Inc.
Burgoyne RD, Weiss JL (2001) The neural calcium-sensor family of Ca2+-binding proteins. Biochem J 353:1–12
Blum JJ, Hayes A, Jamieson GAJ, Vanaman TC (1980) Calmodulin confers calcium sensitivity on ciliary dynein ATPase. J Cell Biol 87:386–397
Tash JS, Krinks M, Patel J, Means RL, Klee CB, Means AR (1988) Identification, characterization, and functional correlation of calmodulin-dependent protein phosphatase in sperm. J Cell Biol 106:1625–1633
Cook SP, Brokaw CJ, Muller CH, Babcock DF (1994) Sperm chemotaxis: egg peptides control cytosolic calcium to regulate flagellar responses. Dev Biol 165:10–19
Eisenbach M (1999) Sperm chemotaxis. Rev Reprod 4:56–66
Kaupp UB, Solzin J, Hildebrand E, Brown JE, Helbig A, Hagen V, Beyermann M, Pampaloni F, Weyand I (2003) The signal flow and motor response controlling chemotaxis of sea urchin sperm. Nat Cell Biol 5:109–117
Wood CD, Nishigaki T, Furuta T, Baba SA, Darszon A (2005) Real-time analysis of the role of Ca2+ in flagellar movement and motility in single sea urchin sperm. J Cell Biol 169:725–731
Miki K, Clapham DE (2013) Rheotaxis guides mammalian sperm. Curr Biol 23:443–452
Kambara Y, Shiba K, Yoshida M, Sato C, Kitajima K, Shingyoji C (2011) Mechanism regulating Ca2+-dependent mechanosensory behaviour in sea urchin spermatozoa. Cell Struct Funct 36:69–82
Saito T, Shiba K, Inaba K, Yamada L, Sawada H (2012) Self-incompatibility response induced by calcium increase in sperm of the ascidian Ciona intestinalis. Proc Natl Acad Sci USA 109:4158–4162
Ishijima S, Mohri H, Overstreet JW, Yudin AI (2006) Hyperactivation of monkey spermatozoa is triggered by Ca2+ and completed by cAMP. Mol Reprod Dev 73:1129–1139
Suarez SS (2008) Control of hyperactivation in sperm. Hum Reprod Update 14:647–657
Chang H, Suarez SS (2010) Rethinking the relationship between hyperactivation and chemotaxis in mammalian sperm. Biol Reprod 83:507–513
Chang H, Suarez SS (2011) Two distinct Ca2+ signaling pathways modulate sperm flagellar beating patterns in mice. Biol Reprod 85:296–305
Yoshida M, Murata M, Inaba K, Morisawa M (2002) A chemoattractant for ascidian spermatozoa is a sulfated steroid. Proc Natl Acad Sci USA 99:14831–14836
Padma P, Satouh Y, Wakabayashi K, Hozumi A, Ushimaru Y, Kamiya R, Inaba K (2003) Identification of a novel leucine-rich repeat protein as a component of flagellar radial spoke in the ascidian Ciona intestinalis. Mol Biol Cell 14:774–785
Mizuno K, Padma P, Konno A, Satouh Y, Ogawa K, Inaba K (2009) A novel neuronal calcium sensor family protein, calaxin, is a potential Ca2+-dependent regulator for the outer arm dynein of metazoan cilia and flagella. Biol Cell 101:91–103
Burgoyne RD (2004) The neuronal calcium-sensor proteins. Biochim Biophys Acta 1742:59–68
Burgoyne RD, O’Callaghan DW, Hasdemir B, Haynes LP, Tepikin AV (2004) Neuronal Ca2+-sensor proteins: multitalented regulators of neuronal function. Trends Neurosci 27:203–209
Tanokura M, Yamada K (1993) A calorimetric study of Ca2+ binding by wheat germ calmodulin. Regulatory steps driven by entropy. J Biol Chem 268:7090–7092
Ames JB, Lim S (2012) Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim Biophysica Acta 1820:1205–1213
Towler DA, Gordon JI, Adams SP, Glaser L (1988) The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem 57:69–99
Sakato M, Sakakibara H, King SM (2007) Chlamydomonas outer arm dynein alters conformation in response to Ca2+. Mol Biol Cell 18:3620–3634
King SM, Patel-King RS (1995) Identification of a Ca2+-binding light chain within Chlamydomonas outer arm dynein. J Cell Sci 108:3757–3764
Cavalier-Smith T (2002) The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52:297–354
Steenkamp ET, Wright J, Baldauf SL (2006) The protistan origins of animals and fungi. Mol Biol Evol 23:93–106
Burki F, Pawlowski J (2006) Monophyly of Rhizaria and multigene phylogeny of unicellular bikonts. Mol Biol Evol 23:1922–1930
Heiss AA, Walker G, Simpson AG (2013) The flagellar apparatus of Breviata anathema, a eukaryote without a clear supergroup affinity. Eur J Protistol 49:354–372
Aldrich HC (1968) The development of flagella in swarm cells of the myxomycete Physarum flavicomum. J Gen Microbiol 50:217–222
Wright M, Tollon Y, Lundström H (1979) Inhibitory action of Nephroma water extracts on the metabolism of Physarum polycephalum. Planta Med 35:323–330
Pagh K, Adelman MR (1982) Identification of a microfilament-enriched, motile domain in amoeboflagellates of Physarum polycephalum. J Cell Sci 54:1–21
Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, Inaba K, Mohri H (1996) Is outer arm dynein intermediate chain 1 multifunctional? Mol Biol Cell 7:1895–907
Padma P, Hozumi A, Ogawa K, Inaba K (2001) Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275:177–183
Satouh Y, Inaba K (2009) Proteomic characterization of sperm radial spokes identifies a novel spoke protein with an ubiquitin domain. FEBS Lett 583:2201–2207
Tang WY, Bell CW, Sale WS, Gibbons IR (1982) Structure of the dynein-1 outer arm in sea urchin sperm flagella. I. Analysis by separation of subunits. J Biol Chem 257:508–515
Yano-Toyoshina Y (1985) Two heavy chains of 21S dynein from sea urchin sperm flagella. J Biochem 98:767–779
Moss AG, Gatti JL, Witman GB (1992) The motile β/IC1 subunit of sea urchin sperm outer arm dynein does not form a rigor bond. J Cell Biol 118:1177–1188
Moss AG, Sale WS, Fox LA, Witman GB (1992) The α subunit of sea urchin sperm outer arm dynein mediates structural and rigor binding to microtubules. J Cell Biol 118:1189–1200
Furuta A, Yagi T, Yanagisawa HA, Higuchi H, Kamiya R (2009) Systematic comparison of in vitro motile properties between Chlamydomonas wild-type and mutant outer arm dyneins each lacking one of the three heavy chains. J Biol Chem 284:5927–5935
Sakato M, King SM (2003) Calcium regulates ATP-sensitive microtubule binding by Chlamydomonas outer arm dynein. J Biol Chem 278:43571–43579
Boitano S, Omoto CK (1991) Membrane hyperpolarization activates trout sperm without an increase in intracellular pH. J Cell Sci 98:343–349
Alavi SM, Gela D, Rodina M, Linhart O (2011) Osmolality, calcium - potassium antagonist and calcium in activation and flagellar beating pattern of sturgeon sperm. Comp Biochem Physiol A 160:166–174
Inouye I, Hori T (1991) High-speed video analysis of the flagellar beat and swimming patterns of algae: possible evolutionary trends in green algae. Protoplasma 164:54–69
Bouck GB (1971) The structure, origin, isolation, and composition of the tubular mastigonemes of the Ochromas flagellum. J Cell Biol 50:362–384
Sleigh MA (1981) Flagellar beat patterns and their possible evolution. Biosystems 14:423–431
Matsunaga S, Uchida H, Iseki M, Watanabe M, Murakami A (2010) Flagellar motions in phototactic steering in a brown algal swarmer. Photochem Photobiol 86:374–381
Takada S, Kamiya R (1994) Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J Cell Biol 126:737–745
Casey DM, Inaba K, Pazour GJ, Takada S, Wakabayashi K, Wilkerson CG, Kamiya R, Witman GB (2003) DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol Biol Cell 14:3650–3663
Hyams JS, Campbell CJ (1985) Widespread absence of outer dynein arms in the spermatozoids of lower plants. Cell Biol Int Rep 9:841–848
Casey DM, Yagi T, Kamiya R, Witman GB (2003) DC3, the smallest subunit of the Chlamydomonas flagellar outer dynein arm-docking complex, is a redox-sensitive calcium-binding protein. J Biol Chem 278:42652–42659
Moestrup Ø, Sengco M (2001) Ultrastructural studies on Bigelowiella natans, gen. et sp. nov., a chlorarachniophyte flagellate. J Phycol 37:624–646
Batistic O, Kudla J (2009) Plant calcineurin B-like proteins and their interacting protein kinases. Biochim Biophys Acta 1793:985–992
Cavalier-Smith T (2006) Cell evolution and earth history: stasis and revolution. Phil Trans R Soc B 361:969–1006
Simpson AG (2003) Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int J Syst Evol Microbiol 53:1759–1777
Andersson JO (2011) Evolution of patchily distributed proteins shared between eukaryotes and prokaryotes: Dictyostelium as a case study. J Mol Microbiol Biotechnol 20:83–95
Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ (2009) Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA 106:3859–3864
Wickstead B, Gull K (2007) Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8:1708–1721
Nicastro D, McIntosh JR, Baumeister W (2005) 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA 102:15889–15894
Ishikawa T, Sakakibara H, Oiwa K (2007) The architecture of outer dynein arms in situ. J Mol Biol 368:1249–1258
Mohri H, Kubo-Irie M, Irie M (1995) Outer arm dynein of sperm flagella and cilia in the animal kingdom. Mém Mus natn Hist nat Paris 166:15–22
Mohri H, Inaba K, Kubo-Irie M, Takai H, Yano-Toyoshima Y (1999) Characterization of outer arm dynein in sea anemone, Anthopleura midori. Cell Motil Cytoskeleton 44:202–208
Mohri H, Inaba K, Ishijima S, Baba SA (2012) Tubulin-dynein system in flagellar and ciliary movement. Proc Jpn Acad Ser B Phys Biol Sci 88:397–415
Marchese-Ragona SP, Gagnon C, White D, Bells Isles H, Johnson KA (1987) Structure and mass analysis of 12S and 19S dynein obtained from bull sperm flagella. Cell Motil Cytoskeleton 8:368–374
Sale WS, Goodeough UW, Heuser JE (1985) The structure of isolated and in situ outer dynein arms of sea urchin sperm flagella. J Cell Biol 101:1400–1412
Sakakibara H, Mitchell DR, Kamiya R (1991) A Chlamydomonas outer dynein mutant missing the α heavy chain. J Cell Biol 113:615–622
Sakakibara H, Takada S, King SM, Witman GB, Kamiya R (1993) A Chlamydomonas outer arm dynein mutant with a truncated β heavy chain. J Cell Biol 122:653–661
Wickstead B, Gull K (2012) Evolutionary biology of dyneins. In: King SM (ed) Dyneins: structure, biology and disease. Academic Press, New York, and Elsevier, Amsterdam, pp 89–121
Tanaka S, Ito S, Kameya-Iwaki M (2001) Electron microscopy of primary zoosporogenesis in Plasmodiophora brassicae. Mycoscience 42:389–394
Straschil U, Talman AM, Ferguson DJ, Bunting KA, Xu Z, Bailes E, Sinden RE, Holder AA, Smith EF, Coates JC, Tewari R (2010) The Armadillo repeat protein PF16 is essential for flagellar structure and function in Plasmodium male gametes. PLoS ONE 5:e12901
Schrevel J, Besse C (1975) A functional flagella with a 6 + 0 pattern. J Cell Biol 66:492–507
Roger AJ, Svard SG, Tovar J, Clark CG, Smith MW, Gillin FD, Sogin ML (1998) Amitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci USA 95:229–234
Friend DS (1966) The fine structure of Giardia muris. J Cell Biol 29:317–332
Embley TM, Hirt RP (1998) Early branching eukaryotes? Curr Opin Genet Dev 8:624–629
Dingle AD, Fulton C (1966) Development of the flagellar apparatus of Naegleria. J Cell Biol 31:43–54
Jemiluhun PF (1998) Isolation and characterization of flagella from Trichomonas vaginalis. Parasitol Res 84:800–805
Cavalier-Smith T (1974) Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci 16:529–556
Mitchell D (2007) The evolution of eukaryotic cilia and flagella as motile and sensory organelles. In: Origins and evolution of eukaryotic endomembranes and cytoskeleton (G. Jekely, ed), Eurekah.com.
Satir P, Guerra C, Bell AJ (2007) Evolution and persistence of the cilium. Cell Motil Cytoskeleton 64:906–913
Satir P, Mitchell DR, Jékely G (2008) How did the cilium evolve? In: Ciliary function in mammalian development. (BK Yoder, ed), Academic Press
Margulis L, Bermudes D (1985) Symbiosis as a mechanism of evolution: status of cell symbiosis theory. Symbiosis 1:101–124
Pickett-Heaps J (1974) The evolution of mitosis and the eukaryotic condition. Biosystem 6:37–48
Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M (2011) Evolution: tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194:165–175
Hodges ME, Wickstead B, Gull K, Langdale JA (2012) The evolution of land plant cilia. New Phytol 195:526–540
Stechmann A, Cavalier-Smith T (2002) Rooting the eukaryote tree by using a derived gene fusion. Science 297:89–91
Richards TA, Cavalier-Smith T (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113–1118
Roger AJ, Simpson AGB (2009) Evolution: revisiting the root of the eukaryote tree. Curr Biol 19:R165–R167
He D, Fiz-Palacios FCJ, Fehling J, Tsai CC, Baldauf SL (2014) An alternative root for the eukaryotic tree of life. Curr Biol 24:465–470
Katz LA, Grant JR, Parfrey LW, Burleigh JG (2012) Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst Biol 61:653–660
Rogozin IB, Basu MK, Csuros M, Koonin EV (2009) Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol 1:99–113
Kamikawa R, Kolisko M, Nishimura Y, Yabuki A, Brown MW, Ishikawa SA, Ishida K, Roger AJ, Hashimoto T, Inagaki Y (2014) Gene content evolution in Discobid mitochondria deduced from the phylogenetic position and complete mitochondrial genome of Tsukubamonas globosa. Genome Biol Evol 6:306–315
Manning G, Reiner DS, Lauwaet T, Dacre M, Smoth A, Zhai Y, Svard S, Gillin FD (2011) The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol 12:R66
Kim E, Simpson AGB, Graham LE (2006) Evolutionary relationships of ausomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol Biol Evol 23:2455–2466
Cavalier-Smith T (2003) Protist phylogeny and the high-level classification of Protozoa. Eur J Protistol 39:338–348
Inaba K (2007) Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann N Y Acad Sci 1101:506–526
Ohno S (1970) Evolution by gene duplication. Springer, New York
Näsvall J, Sun L, Roth JR, Andersson DI (2012) Real-time evolution of new genes by innovation, amplification, and divergence. Science 338:384–387
Ginger ML, Collingridge PW, Nrown RWB, Sproat R, Shaw MK, Gull K (2013) Calmodulin is required for paraflagellar rod assembly and flagellum-cell body attachment in Trypanosomes. Protist 164:528–540
Cavalier-Smith T (1987) The origin of fungi and pseudofungi. In Evolutionary biology of fungi (Rayner ADM,ed.) Cambridge Univ. Press. pp. 339–353
Cotelli F, De Santis R, Rosati F, Monroy A (1980) Acrosome differentiation in the spermatogenesis of Ciona intestinalis. Develop Growth Differ 22:561–569
Moreno RD, Ramalho-Santos J, Sutovsky P, Chan EK, Schatten G (2000) Vesicular traffic and Golgi apparatus dynamics during mammalian spermatogenesis: implications for acrosome architecture. Biol Reprod 63:89–98
Goodenough UW, Detmers PA, Hwang C (1982) Activation for cell fusion in Chlamydomonas: analysis of wild-type gametes and nonfusing mutants. J Cell Biol 92:378–386
Inaba K (2012) Regulatory subunits of axonemal dynein. In: Handbook of dynein (eds. Hirose, Amos). PanStanford Publishing, pp. 303–324
Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–D392
Bouck GB, Rogalski A, Valaitis A (1978) Surface organization and composition of Euglena. II. Flagellar mastigonemes. J Cell Biol 77:805
Suzaki T, Williamson RE (1986) Reactivation of euglenoid movement and flagellar beating in detergent-extracted cells of Astasia longa: different mechanisms of force generation are involved. J Cell Sci 80:75–89
Hibberd DJ (1975) Observation on the ultrastructure of the choanoflagellate Codosiga botrytis (HER.) Saville-Kent with special reference to the flagellar apparatus. J Cell Sci 17:191–219
McKanna JA (1968) Fine structure of the protonephridial system in Planaria. I. Flame cells. Z Zellforsch Mikrosk Anat 92:509–523
Dallai R, Bellon PL, Lanzavecchia S, Afzelius BA (1993) The dipteran sperm tail: ultrastructural characteristics and phylogenetic considerations. Zoologica Scripta 22:193–202
Wada S, Okuno M, Nakamura K, Mohri H (1992) Dynein of sperm flagella of oyster beloning to protostomia also has a two-headed structure. Biol Cell 76:311–317
Barr DJS, Désaulniers ND (1986) Four zoospore subtypes in the Rhizophlyctis-Karlingia complex (Chytridiomycetes). Can J Bot 64:561–572
Heath B, Darley WM (1972) Observations on the ultrastructure of the male gametes of Biddulphia levis ehr.1. J Phycol 8:51–59
Allen RD (1968) A reinvestigation of cross-sections of cilia. J Cell Biol 37:825–831
Leadbeater B, Dodge JD (1967) An electron microscope study of dinoflagellate flagella. J Gen Microbiol 46:305–314
Birkhead M, Pienaar RN (1995) The flagellar apparatus of Chrysochromulina sp. (Prymnesiophyceae). J Phycol 31:96–108
Acknowledgements
This work was supported in part by Grant-in-aid No. 21112004 for Scientific Research on Innovative Areas, No. 16083203 for Scientific Research on Priority Areas, and No. 22370023 for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). Thanks are also due to all staff members of the Education and Research Center of Marine Bio-Resources (Onagawa), Tohoku University, International Coastal Research Center (Otsuchi), The University of Tokyo, and JAMBIO (Japanese Association for Marine Biology) who supported this work. I am indebted to Dr. Zac Cande and his colleagues in helping me to find the TEM image of Naegleria axonemes. I would like to acknowledge all the papers with TEM images of axonemes used for analysis, listed in the legend of Figure 9.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author declares that he has no competing interests.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Inaba, K. Calcium sensors of ciliary outer arm dynein: functions and phylogenetic considerations for eukaryotic evolution. Cilia 4, 6 (2015). https://doi.org/10.1186/s13630-015-0015-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13630-015-0015-z